1RTN
 
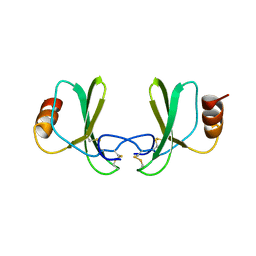 | |
1RTO
 
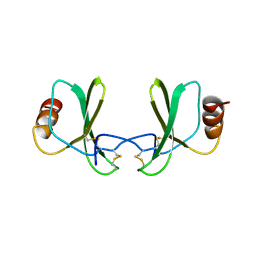 | |
1CLB
 
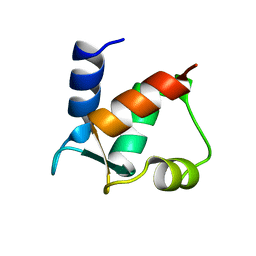 | |
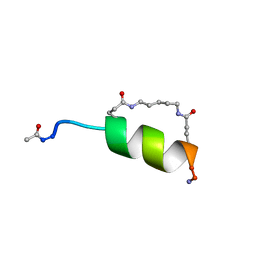
1IN2
 
 | | Peptide Antagonist of IGFBP1, (i,i+7) Covalently Restrained Analog | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1IN3
 
 | | Peptide Antagonist of IGFBP1, (i,i+8) Covalently Restrained Analog | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1GJF
 
 | | Peptide Antagonist of IGFBP1, (i,i+7) Covalently Restrained Analog, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1GJG
 
 | | Peptide Antagonist of IGFBP1, (i,i+8) Covalently Restrained Analog, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1ILP
 
 | |
1KVG
 
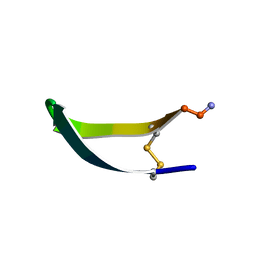 | | EPO-3 beta Hairpin Peptide | | Descriptor: | Protein: EPO-3 Receptor Agonist | | Authors: | Skelton, N.J, Russell, S, de Sauvage, F, Cochran, A.G. | | Deposit date: | 2002-01-25 | | Release date: | 2002-03-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Amino Acid Determinants of beta-Hairpin Conformation in Erythropoeitin
Receptor Agonist Peptides Derived from a Phage Display Library
J.Mol.Biol., 316, 2002
|
|
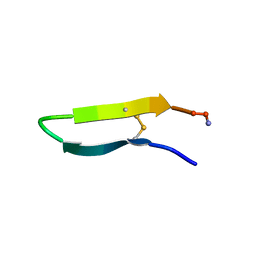
1KVF
 
 | |
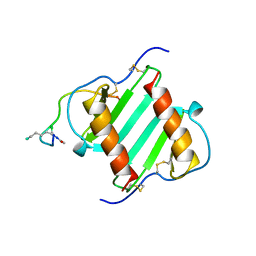
1ILQ
 
 | |
1GNB
 
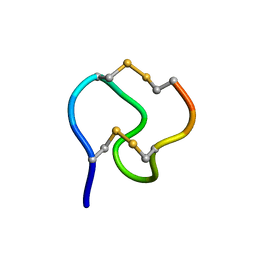 | |
1GNA
 
 | |
1QSZ
 
 | | THE VEGF-BINDING DOMAIN OF FLT-1 (MINIMIZED MEAN) | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 1 | | Authors: | Starovasnik, M.A, Christinger, H.W, Wiesmann, C, Champe, M.A, de Vos, A.M, Skelton, N.J. | | Deposit date: | 1999-06-24 | | Release date: | 1999-11-10 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the VEGF-binding domain of Flt-1: comparison of its free and bound states.
J.Mol.Biol., 293, 1999
|
|
4N9D
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, 4-({[(4-tert-butylphenyl)sulfonyl]amino}methyl)-N-(pyridin-3-yl)benzamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9B
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1-methyl-N-(pyridin-3-yl)-1H-pyrazole-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhai, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.859 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9E
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1-benzoylpiperidin-4-yl)methyl]-N-(pyridin-3-yl)-1H-benzimidazole-5-carboxamide, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2BCA
 
 | |
2BCB
 
 | |
4N9C
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 5-nitro-1H-benzimidazole, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1XUT
 
 | | Solution structure of TACI-CRD2 | | Descriptor: | Tumor necrosis factor receptor superfamily member 13B | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structures of APRIL-receptor complexes: like BCMA, TACI employs only a single cysteine-rich domain for high affinity ligand binding.
J.Biol.Chem., 280, 2005
|
|
1XU1
 
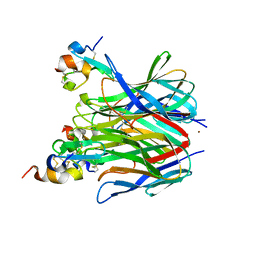 | | The crystal structure of APRIL bound to TACI | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 13B | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
1XU2
 
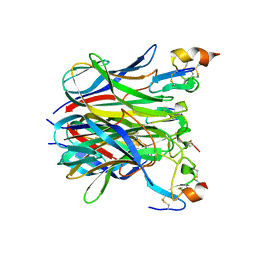 | | The crystal structure of APRIL bound to BCMA | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
3QKL
 
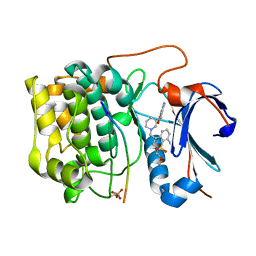 | | Spirochromane Akt Inhibitors | | Descriptor: | GSK-3 beta peptide, N-{(2S)-3-[(3S)-8',9'-dihydro-1H,3'H-spiro[piperidine-3,7'-pyrano[3,2-e]indazol]-1-yl]-2-hydroxypropyl}-N-(2-ethoxyethyl)-2,6-dimethylbenzenesulfonamide, RAC-alpha serine/threonine-protein kinase | | Authors: | Kallan, N.C, Spencer, K.L, Blake, J.F, Xu, R, Heizer, J, Bencsik, J.R, Mitchell, I.S, Gloor, S.L, Martinson, M, Risom, T, Gross, S.D, Morales, T, Vigers, G.P.A, Brandhuber, B.J, Skelton, N.J. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and SAR of spirochromane Akt inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2JOA
 
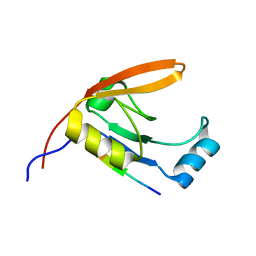 | | HtrA1 bound to an optimized peptide: NMR assignment of PDZ domain and ligand resonances | | Descriptor: | Peptide H1-C1, Serine protease HTRA1 | | Authors: | Runyon, S.T, Zhang, Y, Appleton, B.A, Sazinksy, S.L, Wu, P, Pan, B, Wiesmann, C, Skelton, N.J, Sidhu, S.S. | | Deposit date: | 2007-03-01 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and functional analysis of the PDZ domains of human HtrA1 and HtrA3
Protein Sci., 16, 2007
|
|
