2B0V
 
 | | NUDIX hydrolase from Nitrosomonas europaea. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NUDIX hydrolase, ... | | Authors: | Osipiuk, J, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-14 | | Release date: | 2005-09-27 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure of NUDIX hydrolase from Nitrosomonas europaea.
To be Published
|
|
2AUW
 
 | | Crystal Structure of Putative DNA Binding Protein NE0471 from Nitrosomonas europaea ATCC 19718 | | Descriptor: | FORMIC ACID, GLYCEROL, hypothetical protein NE0471 | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-29 | | Release date: | 2005-10-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Hypothetical Protein NE0471 from Nitrosomonas europaea
To be Published
|
|
2FA1
 
 | | Crystal structure of PhnF C-terminal domain | | Descriptor: | Probable transcriptional regulator phnF, beta-D-fructopyranose | | Authors: | Lunin, V.V, Nocek, B.P, Gorelik, M, Skarina, T, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-06 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of GntR/HutC family signaling domain.
Protein Sci., 15, 2006
|
|
2ESN
 
 | | The crystal structure of probable transcriptional regulator PA0477 from Pseudomonas aeruginosa | | Descriptor: | probable transcriptional regulator | | Authors: | Lunin, V.V, Chang, C, Skarina, T, Gorodischenskaya, E, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of putative transcriptional regulator Pa0477 from Pseudomonas aeruginosa
To be Published
|
|
2G8Y
 
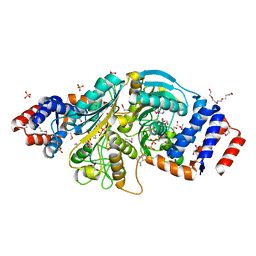 | | The structure of a putative malate/lactate dehydrogenase from E. coli. | | Descriptor: | 1,2-ETHANEDIOL, Malate/L-lactate dehydrogenases, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of a putative malate/lactate dehydrogenase from E. coli.
To be Published
|
|
2FD5
 
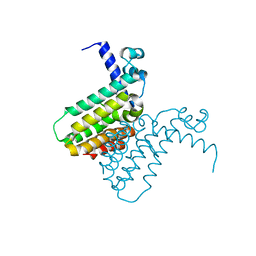 | | The crystal structure of a transcriptional regulator from Pseudomonas aeruginosa PAO1 | | Descriptor: | transcriptional regulator | | Authors: | Zhang, R, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-13 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a transcriptional regulator from Pseudomonas aeruginosa PAO1
To be Published
|
|
2FIW
 
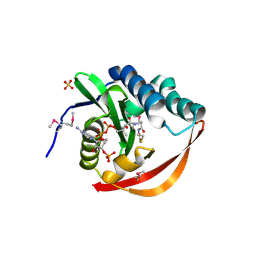 | | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris | | Descriptor: | ACETYL COENZYME *A, GCN5-related N-acetyltransferase:Aminotransferase, class-II, ... | | Authors: | Kim, Y, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-30 | | Release date: | 2006-02-14 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris
To be Published, 2006
|
|
2O9X
 
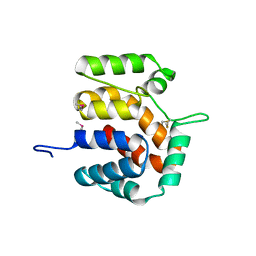 | | Crystal Structure Of A Putative Redox Enzyme Maturation Protein From Archaeoglobus Fulgidus | | Descriptor: | Reductase, assembly protein | | Authors: | Kirillova, O, Chruszcz, M, Skarina, T, Gorodichtchenskaia, E, Cymborowski, M, Shumilin, I, Savchenko, A, Edwards, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-14 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An extremely SAD case: structure of a putative redox-enzyme maturation protein from Archaeoglobus fulgidus at 3.4 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2NS0
 
 | | Crystal structure of protein RHA04536 from Rhodococcus sp | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hypothetical protein | | Authors: | Chang, C, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-02 | | Release date: | 2006-12-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal structure of protein RHA04536 from Rhodococcus sp
To be Published
|
|
2ODK
 
 | | Putative prevent-host-death protein from Nitrosomonas europaea | | Descriptor: | GLYCEROL, Hypothetical protein, SULFATE ION | | Authors: | Osipiuk, J, Skarina, T, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-22 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crsytal structure of putative prevent-host-death protein from Nitrosomonas europaea.
To be Published
|
|
2OER
 
 | | Probable Transcriptional Regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator | | Authors: | Kim, Y, Skarina, T, Kagan, O, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-31 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Probable Transcriptional Regulator from Pseudomonas aeruginosa
To be Published
|
|
2OFY
 
 | | Crystal structure of putative XRE-family transcriptional regulator from Rhodococcus sp. | | Descriptor: | Putative XRE-family transcriptional regulator | | Authors: | Shumilin, I.A, Skarina, T, Onopriyenko, O, Yim, V, Chruszcz, M, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-04 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative XRE-family transcriptional regulator from Rhodococcus sp.
To be Published
|
|
2O0Y
 
 | | Crystal structure of putative transcriptional regulator RHA1_ro06953 (IclR-family) from Rhodococcus sp. | | Descriptor: | Transcriptional regulator | | Authors: | Chruszcz, M, Wang, S, Skarina, T, Onopriyenko, O, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Putative Transcriptional Regul 2 Rha1_Ro06953(Iclr-Family) from Rhodococcus Sp.
To be Published
|
|
1U60
 
 | | MCSG APC5046 Probable glutaminase ybaS | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable glutaminase ybaS | | Authors: | Chang, C, Cuff, M.E, Joachimiak, A, Savchenko, A, Edwards, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
2PJS
 
 | | Crystal structure of Atu1953, protein of unknown function | | Descriptor: | Uncharacterized protein Atu1953, ZINC ION | | Authors: | Chang, C, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Glyoxalase/Bleomycin resistance protein/Dioxygenase superfamily protein Atu1953, protein of unknown function
To be Published
|
|
2PYU
 
 | | Structure of the E. coli inosine triphosphate pyrophosphatase RgdB in complex with IMP | | Descriptor: | 1,2-ETHANEDIOL, INOSINIC ACID, Inosine Triphosphate Pyrophosphatase RdgB | | Authors: | Singer, A.U, Proudfoot, M, Skarina, T, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2007-05-16 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
2OR0
 
 | | Structural Genomics, the crystal structure of a putative hydroxylase from Rhodococcus sp. RHA1 | | Descriptor: | ACETATE ION, Hydroxylase | | Authors: | Tan, K, Skarina, T, Kagen, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-01 | | Release date: | 2007-03-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a putative hydroxylase from Rhodococcus sp. RHA1
To be Published
|
|
2PPX
 
 | | Crystal structure of a HTH XRE-family like protein from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, SULFATE ION, Uncharacterized protein Atu1735 | | Authors: | Cuff, M.E, Skarina, T, Onopriyenko, O, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-30 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a HTH XRE-family like protein from Agrobacterium tumefaciens.
TO BE PUBLISHED
|
|
2Q05
 
 | | Crystal structure of tyr/ser protein phosphatase from Vaccinia virus WR | | Descriptor: | Dual specificity protein phosphatase | | Authors: | Osipiuk, J, Skarina, T, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-18 | | Release date: | 2007-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of tyr/ser protein phosphatase from Vaccinia virus WR.
To be Published
|
|
2Q16
 
 | | Structure of the E. coli inosine triphosphate pyrophosphatase RgdB in complex with ITP | | Descriptor: | CALCIUM ION, HAM1 protein homolog, INOSINE 5'-TRIPHOSPHATE, ... | | Authors: | Singer, A.U, Lam, R, Proudfoot, M, Skarina, T, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2007-05-23 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
8EJU
 
 | | The crystal structure of Pseudomonas putida PcaR | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Transcription regulatory protein (Pca regulon), ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The crystal structure of Pseudomonas putida PcaR
To Be Published
|
|
1SDJ
 
 | | X-RAY STRUCTURE OF YDDE_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET25. | | Descriptor: | Hypothetical protein yddE, SULFATE ION | | Authors: | Kuzin, A.P, Edstrom, W, Skarina, T, Korniyenko, Y, Savchenko, A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
2GEN
 
 | | Structural Genomics, the crystal structure of a probable transcriptional regulator from Pseudomonas aeruginosa PAO1 | | Descriptor: | probable transcriptional regulator | | Authors: | Tan, K, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-20 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a probable transcriptional regulator from Pseudomonas aeruginosa PAO1
To be Published
|
|
3EEF
 
 | | Crystal structure of N-carbamoylsarcosine amidase from thermoplasma acidophilum | | Descriptor: | N-carbamoylsarcosine amidase related protein, ZINC ION | | Authors: | Luo, H.-B, Zheng, H, Chruszcz, M, Zimmerman, M.D, Skarina, T, Egorova, O, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and molecular modeling study of N-carbamoylsarcosine amidase Ta0454 from Thermoplasma acidophilum.
J.Struct.Biol., 169, 2010
|
|
3E4F
 
 | | Crystal structure of BA2930- a putative aminoglycoside N3-acetyltransferase from Bacillus anthracis | | Descriptor: | Aminoglycoside N3-acetyltransferase, CITRIC ACID | | Authors: | Klimecka, M.M, Chruszcz, M, Skarina, T, Onopryienko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-11 | | Release date: | 2008-08-19 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
