4AYC
 
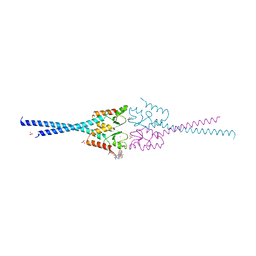 | | RNF8 RING domain structure | | Descriptor: | CHLORIDE ION, E3 UBIQUITIN-PROTEIN LIGASE RNF8, GLYCEROL, ... | | Authors: | Mattiroli, F, Vissers, J.H.A, Van Dijk, W.J, Ikpa, P, Citterio, E, Vermeulen, W, Marteijn, J.A, Sixma, T.K. | | Deposit date: | 2012-06-20 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rnf168 Ubiquitinates K13-15 on H2A/H2Ax to Drive DNA Damage Signaling
Cell(Cambridge,Mass.), 150, 2012
|
|
1U9A
 
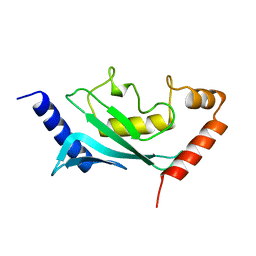 | | HUMAN UBIQUITIN-CONJUGATING ENZYME UBC9 | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME | | Authors: | Tong, H, Hateboer, G, Perrakis, A, Bernards, R, Sixma, T.K. | | Deposit date: | 1997-02-11 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of murine/human Ubc9 provides insight into the variability of the ubiquitin-conjugating system.
J.Biol.Chem., 272, 1997
|
|
2BR8
 
 | | Crystal Structure of Acetylcholine-binding Protein (AChBP) from Aplysia californica in complex with an alpha-conotoxin PnIA variant | | Descriptor: | ALPHA-CONOTOXIN PNIA, SOLUBLE ACETYLCHOLINE RECEPTOR, SULFATE ION | | Authors: | Celie, P.H.N, Kasheverov, I.E, Mordvintsev, D.Y, Hogg, R.C, van Nierop, P, van Elk, R, van Rossum-Fikkert, S.E, Zhmak, M.N, Bertrand, D, Tsetlin, V, Sixma, T.K, Smit, A.B. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Nicotinic Acetylcholine Receptor Homolog Achbp in Complex with an Alpha-Conotoxin Pnia Variant
Nat.Struct.Mol.Biol., 12, 2005
|
|
1WB9
 
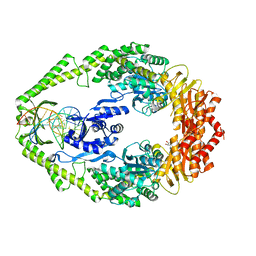 | | Crystal Structure of E. coli DNA Mismatch Repair enzyme MutS, E38T mutant, in complex with a G.T mismatch | | Descriptor: | 5'-D(*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Georgijevic, D, Lebbink, J.H.G, Winterwerp, H.H.K, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-10-31 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Embo J., 25, 2006
|
|
1WBB
 
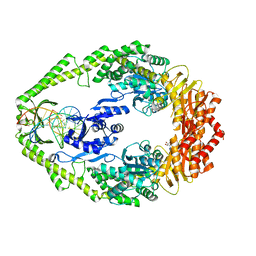 | | Crystal structure of E. coli DNA mismatch repair enzyme MutS, E38A mutant, in complex with a G.T mismatch | | Descriptor: | 5'-D(*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Georgijevic, D, Lebbink, J.H.G, Winterwerp, H.H.K, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-10-31 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Embo J., 25, 2006
|
|
1WBD
 
 | | Crystal structure of E. coli DNA mismatch repair enzyme MutS, E38Q mutant, in complex with a G.T mismatch | | Descriptor: | 5'-D(*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Georgijevic, D, Lebbink, J.H.G, Winterwerp, H.H.K, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-10-31 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Embo J., 25, 2006
|
|
3ZLJ
 
 | | CRYSTAL STRUCTURE OF FULL-LENGTH E.COLI DNA MISMATCH REPAIR PROTEIN MUTS D835R MUTANT IN COMPLEX WITH GT MISMATCHED DNA | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP *AP*GP*TP*GP*TP*CP*AP)-3', 5'-D(*TP*GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*TP)-3', DNA MISMATCH REPAIR PROTEIN MUTS | | Authors: | Groothuizen, F.S, Fish, A, Petoukhov, M.V, Reumer, A, Manelyte, L, Winterwerp, H.H.K, Marinus, M.G, Lebbink, J.H.G, Svergun, D.I, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2013-02-01 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Using Stable Muts Dimers and Tetramers to Quantitatively Analyze DNA Mismatch Recognition and Sliding Clamp Formation.
Nucleic Acids Res., 41, 2013
|
|
1W7A
 
 | | ATP bound MutS | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP* AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP* CP*T)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lamers, M.H, Georgijevic, D, Lebbink, J, Winterwerp, H.H.K, Agianian, B, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-08-31 | | Release date: | 2004-09-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | ATP Increases the Affinity between Muts ATPase Domains: Implications for ATP Hydrolysis and Conformational Changes
J.Biol.Chem., 279, 2004
|
|
1NG9
 
 | | E.coli MutS R697A: an ATPase-asymmetry mutant | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lamers, M.H, Winterwerp, H.H.K, Sixma, T.K. | | Deposit date: | 2002-12-17 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The alternating ATPase domains of MutS control DNA mismatch repair
Embo J., 22, 2003
|
|
2BF8
 
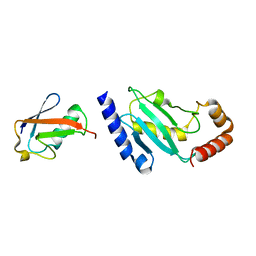 | | Crystal structure of SUMO modified ubiquitin conjugating enzyme E2- 25K | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2-25 KDA, UBIQUITIN-LIKE PROTEIN SMT3C | | Authors: | Pichler, A, Knipscheer, P, Oberhofer, E, Van Dijk, W.J, Korner, R, Velgaard Olsen, J, Jentsch, S, Melchior, F, Sixma, T.K. | | Deposit date: | 2004-12-06 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sumo Imodification of the Ubiquitin Conjugating Enzyme E2-25K
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BEP
 
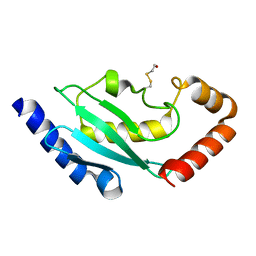 | | Crystal structure of ubiquitin conjugating enzyme E2-25K | | Descriptor: | BETA-MERCAPTOETHANOL, UBIQUITIN-CONJUGATING ENZYME E2-25 KDA | | Authors: | Pichler, A, Knipscheer, P, Oberhofer, E, Van Dijk, W.J, Korner, R, Velgaard Olsen, J, Jentsch, S, Melchior, F, Sixma, T.K. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sumo Imodification of the Ubiquitin Conjugating Enzyme E2-25K
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BR7
 
 | | Crystal Structure of Acetylcholine-binding Protein (AChBP) from Aplysia californica in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Celie, P.H.N, Kasheverov, I.E, Mordvintsev, D.Y, Hogg, R.C, Van Nierop, P, Van Elk, R, Van Rossum-Fikkert, S.E, Zhmak, M.N, Bertrand, D, Tsetlin, V, Sixma, T.K, Smit, A.B. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Nicotinic Acetylcholine Receptor Homolog Achbp in Complex with an Alpha- Conotoxin Pnia Variant
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BJ0
 
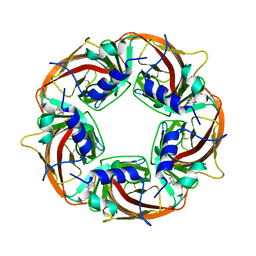 | | Crystal Structure of AChBP from Bulinus truncatus revals the conserved structural scaffold and sites of variation in nicotinic acetylcholine receptors | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Celie, P.H.N, Klaassen, R.V, Van Rossum-Fikkert, S.E, Van Elk, R, Van Nierop, P, Smit, A.B, Sixma, T.K. | | Deposit date: | 2005-01-27 | | Release date: | 2005-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Acetylcholine-Binding Protein from Bulinus Truncatus Reveals the Conserved Structural Scaffold and Sites of Variation in Nicotinic Acetylcholine Receptors.
J.Biol.Chem., 280, 2005
|
|
2Y6E
 
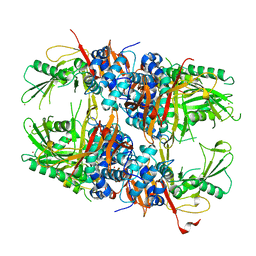 | | Structure of the D1D2 domain of USP4, the conserved catalytic domain | | Descriptor: | SULFATE ION, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 4, ZINC ION | | Authors: | Luna-Vargas, M.P.A, Faesen, A.C, van Dijk, W.J, Rape, M, Fish, A, Sixma, T.K. | | Deposit date: | 2011-01-20 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Dusp-Ubl Domain of Usp4 Enhances its Catalytic Efficiency by Promoting Ubiquitin Exchange.
Nat.Commun., 5, 2014
|
|
2YLM
 
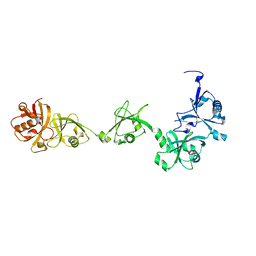 | |
2Y58
 
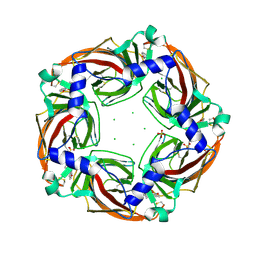 | | Fragment growing induces conformational changes in acetylcholine- binding protein: A structural and thermodynamic analysis - (Compound 6) | | Descriptor: | CHLORIDE ION, SOLUBLE ACETYLCHOLINE RECEPTOR, SULFATE ION, ... | | Authors: | Rucktooa, P, Edink, E, deEsch, I.J.P, Sixma, T.K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Fragment Growing Induces Conformational Changes in Acetylcholine-Binding Protein: A Structural and Thermodynamic Analysis.
J.Am.Chem.Soc., 133, 2011
|
|
2Y54
 
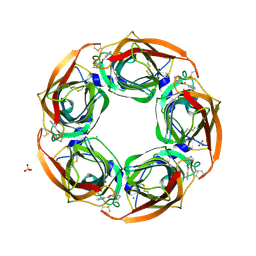 | | Fragment growing induces conformational changes in acetylcholine- binding protein: A structural and thermodynamic analysis - (Fragment 1) | | Descriptor: | CHLORIDE ION, SOLUBLE ACETYLCHOLINE RECEPTOR, SULFATE ION, ... | | Authors: | Rucktooa, P, Edink, E, deEsch, I.J.P, Sixma, T.K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Fragment Growing Induces Conformational Changes in Acetylcholine-Binding Protein: A Structural and Thermodynamic Analysis.
J.Am.Chem.Soc., 133, 2011
|
|
2Y56
 
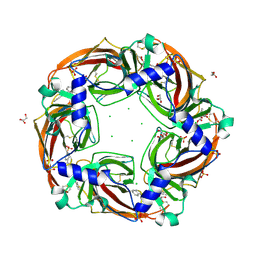 | | Fragment growing induces conformational changes in acetylcholine- binding protein: A structural and thermodynamic analysis - (Compound 3) | | Descriptor: | CHLORIDE ION, GLYCEROL, SOLUBLE ACETYLCHOLINE RECEPTOR, ... | | Authors: | Rucktooa, P, Edink, E, deEsch, I.J.P, Sixma, T.K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Fragment Growing Induces Conformational Changes in Acetylcholine-Binding Protein: A Structural and Thermodynamic Analysis.
J.Am.Chem.Soc., 133, 2011
|
|
2Y57
 
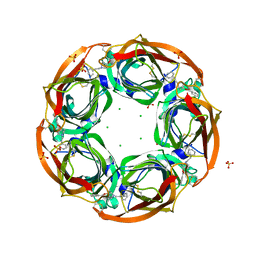 | | Fragment growing induces conformational changes in acetylcholine- binding protein: A structural and thermodynamic analysis - (Compound 4) | | Descriptor: | CHLORIDE ION, SOLUBLE ACETYLCHOLINE RECEPTOR, SULFATE ION, ... | | Authors: | Rucktooa, P, Edink, E, deEsch, I.J.P, Sixma, T.K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fragment Growing Induces Conformational Changes in Acetylcholine-Binding Protein: A Structural and Thermodynamic Analysis.
J.Am.Chem.Soc., 133, 2011
|
|
2XNU
 
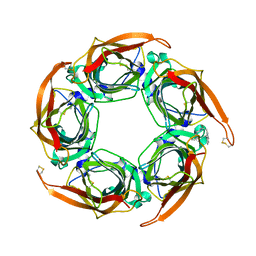 | | Acetylcholine binding protein (AChBP) as template for hierarchical in silico screening procedures to identify structurally novel ligands for the nicotinic receptors | | Descriptor: | 2-(2-(4-PHENYLPIPERIDIN-1-YL)ETHYL)-1H-INDOLE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Rucktooa, P, Akdemir, A, deEsch, I, Sixma, T.K. | | Deposit date: | 2010-08-06 | | Release date: | 2011-08-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Acetylcholine Binding Protein (Achbp) as Template for Hierarchical in Silico Screening Procedures to Identify Structurally Novel Ligands for the Nicotinic Receptors.
Bioorg.Med.Chem., 19, 2011
|
|
2XNT
 
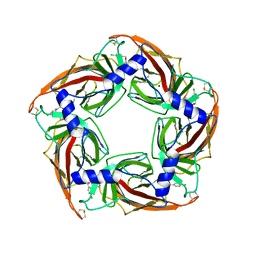 | | Acetylcholine binding protein (AChBP) as template for hierarchical in silico screening procedures to identify structurally novel ligands for the nicotinic receptors | | Descriptor: | (2S)-2-[(4-CHLOROBENZYL)OXY]-2-PHENYLETHANAMINE, BROMIDE ION, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Rucktooa, P, Akdemir, A, deEsch, I, Sixma, T.K. | | Deposit date: | 2010-08-06 | | Release date: | 2011-08-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Acetylcholine Binding Protein (Achbp) as Template for Hierarchical in Silico Screening Procedures to Identify Structurally Novel Ligands for the Nicotinic Receptors.
Bioorg.Med.Chem., 19, 2011
|
|
2XNV
 
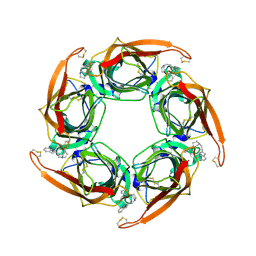 | | Acetylcholine binding protein (AChBP) as template for hierarchical in silico screening procedures to identify structurally novel ligands for the nicotinic receptors | | Descriptor: | 2-(2-(4-PHENYLPIPERIDIN-1-YL)ETHYL)-1H-INDOLE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Rucktooa, P, Akdemir, A, deEsch, I, Sixma, T.K. | | Deposit date: | 2010-08-06 | | Release date: | 2011-08-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Acetylcholine Binding Protein (Achbp) as Template for Hierarchical in Silico Screening Procedures to Identify Structurally Novel Ligands for the Nicotinic Receptors.
Bioorg.Med.Chem., 19, 2011
|
|
1U78
 
 | |
2WTU
 
 | | Crystal structure of Escherichia coli MutS in complex with a 16 basepair oligo containing an A.A mismatch. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, DNA, ... | | Authors: | Natrajan, G, Lebbink, J.H, Reumer, A, Fish, A, Winterwerp, H.H, Sixma, T.K. | | Deposit date: | 2009-09-22 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Magnesium coordination controls the molecular switch function of DNA mismatch repair protein MutS.
J. Biol. Chem., 285, 2010
|
|
2W8F
 
 | | Aplysia californica AChBP bound to in silico compound 31 | | Descriptor: | (3-EXO)-3-(10,11-DIHYDRO-5H-DIBENZO[A,D][7]ANNULEN-5-YLOXY)-8,8-DIMETHYL-8-AZONIABICYCLO[3.2.1]OCTANE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Akdemir, A, Jongejan, A, van Elk, R, Edink, E, Bertrand, S, Perrakis, A, Leurs, R, Smit, A.B, Sixma, T.K, Bertrand, D, de Esch, I.J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Use of Acetylcholine Binding Protein in the Search for Novel Alpha7 Nicotinic Receptor Ligands. In Silico Docking, Pharmacological Screening, and X- Ray Analysis.
J.Med.Chem., 52, 2009
|
|
