2ZQS
 
 | |
2ZQV
 
 | |
2ZQU
 
 | |
2ZR4
 
 | |
2ZR5
 
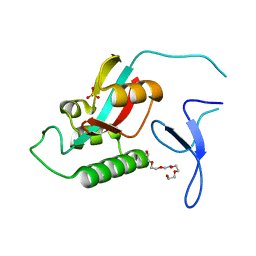 | |
2GJ2
 
 | | Crystal Structure of VP9 from White Spot Syndrome Virus | | Descriptor: | CADMIUM ION, wsv230 | | Authors: | Liu, Y, Wu, J.L, Song, J.X, Sivaraman, J, Hew, C.L. | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of a Novel Nonstructural Protein, VP9, from White Spot Syndrome Virus: Its Structure Reveals a Ferredoxin Fold with Specific Metal Binding Sites
J.Virol., 80, 2006
|
|
2GJI
 
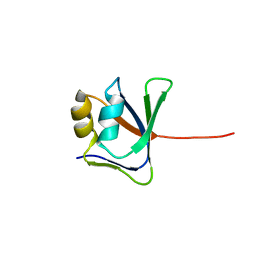 | | NMR solution structure of VP9 from White Spot Syndrome Virus | | Descriptor: | wsv230 | | Authors: | Liu, Y, Wu, J.L, Song, J.X, Sivaraman, J, Hew, C.L. | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Identification of a Novel Nonstructural Protein, VP9, from White Spot Syndrome Virus: Its Structure Reveals a Ferredoxin Fold with Specific Metal Binding Sites
J.Virol., 80, 2006
|
|
2PJD
 
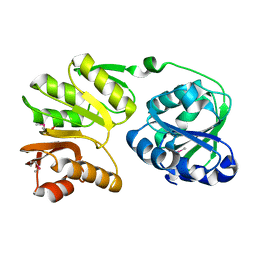 | | Crystal structure of 16S rRNA methyltransferase RsmC | | Descriptor: | Ribosomal RNA small subunit methyltransferase C | | Authors: | Sunita, S, Purta, E, Durawa, M, Tkaczuk, K.L, Bujnicki, J.M, Sivaraman, J. | | Deposit date: | 2007-04-16 | | Release date: | 2007-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional specialization of domains tandemly duplicated within 16S rRNA methyltransferase RsmC
Nucleic Acids Res., 35, 2007
|
|
2MQ1
 
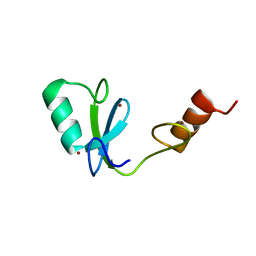 | | Phosphotyrosine binding domain | | Descriptor: | E3 ubiquitin-protein ligase Hakai, ZINC ION | | Authors: | Mukherjee, M, Jing-Song, F, Sivaraman, J. | | Deposit date: | 2014-06-11 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dimeric switch of Hakai-truncated monomers during substrate recognition: insights from solution studies and NMR structure.
J.Biol.Chem., 289, 2014
|
|
1EOJ
 
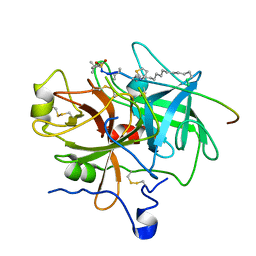 | | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures | | Descriptor: | ALPHA THROMBIN, THROMBIN INHIBITOR P798 | | Authors: | Slon-Usakiewicz, J.J, Sivaraman, J, Li, Y, Cygler, M, Konishi, Y. | | Deposit date: | 2000-03-23 | | Release date: | 2000-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures.
Biochemistry, 39, 2000
|
|
5E38
 
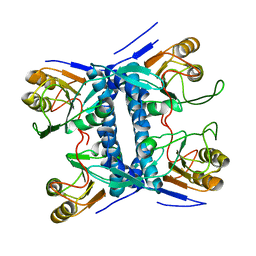 | | Structural basis of mapping the spontaneous mutations with 5-flourouracil in uracil phosphoribosyltransferase from Mycobacterium tuberculosis | | Descriptor: | Uracil phosphoribosyltransferase | | Authors: | Ghode, P, Jobichen, C, Ramachandran, S, Bifani, P, Sivaraman, J. | | Deposit date: | 2015-10-02 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of mapping the spontaneous mutations with 5-flurouracil in uracil phosphoribosyltransferase from Mycobacterium tuberculosis
Biochem.Biophys.Res.Commun., 467, 2015
|
|
1EOL
 
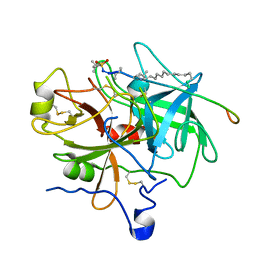 | | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures | | Descriptor: | ALPHA THROMBIN, THROMBIN INHIBITOR P628 | | Authors: | Slon-Usakiewicz, J.J, Sivaraman, J, Li, Y, Cygler, M, Konishi, Y. | | Deposit date: | 2000-03-23 | | Release date: | 2000-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures.
Biochemistry, 39, 2000
|
|
1FC4
 
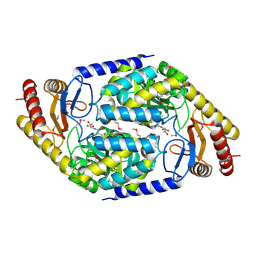 | | 2-AMINO-3-KETOBUTYRATE COA LIGASE | | Descriptor: | 2-AMINO-3-KETOBUTYRATE CONENZYME A LIGASE, 2-AMINO-3-KETOBUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schmidt, A, Matte, A, Li, Y, Sivaraman, J, Larocque, R, Schrag, J.D, Smith, C, Sauve, V, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2000-07-17 | | Release date: | 2001-05-02 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of 2-amino-3-ketobutyrate CoA ligase from Escherichia coli complexed with a PLP-substrate intermediate: inferred reaction mechanism.
Biochemistry, 40, 2001
|
|
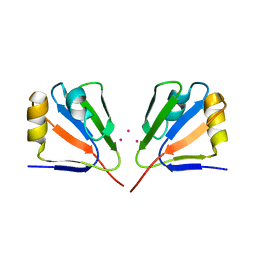
3GGQ
 
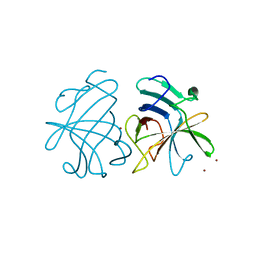 | | Dimerization of Hepatitis E Virus Capsid Protein E2s Domain is Essential for Virus-Host Interaction | | Descriptor: | BROMIDE ION, Capsid protein | | Authors: | Li, S.W, Tang, X.H, Seetharaman, J, Yang, C.Y, Gu, Y, Zhang, J, Du, H.L, Shih, J.W.K, Hew, C.L, Sivaraman, J, Xia, N.S. | | Deposit date: | 2009-03-02 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dimerization of hepatitis E virus capsid protein E2s domain is essential for virus-host interaction
Plos Pathog., 5, 2009
|
|
4HEX
 
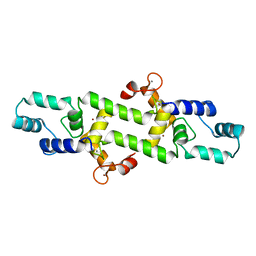 | |
5XWE
 
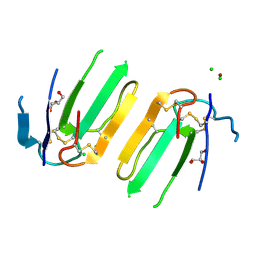 | | Structure of a three finger toxin from Ophiophagus hannah venom | | Descriptor: | CHLORIDE ION, GLYCEROL, Weak toxin DE-1 homolog 1, ... | | Authors: | Jobichen, C, Roy, A, Kini, R.M, Sivaraman, J. | | Deposit date: | 2017-06-29 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a three finger toxin from Ophiophagus hannah venom
To Be Published
|
|
6K3O
 
 | |
6K4M
 
 | |
6K43
 
 | |
5YDX
 
 | |
5YDY
 
 | |
1MHW
 
 | | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides | | Descriptor: | 4-biphenylacetyl-Cys-(D)Arg-Tyr-N-(2-phenylethyl) amide, Cathepsin L | | Authors: | Chowdhury, S, Sivaraman, J, Wang, J, Devanathan, G, Lachance, P, Qi, H, Menard, R, Lefebvre, J, Konishi, Y, Cygler, M, Sulea, T, Purisima, E.O. | | Deposit date: | 2002-08-21 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides
J.Med.Chem., 45, 2002
|
|
4KT5
 
 | |
7C28
 
 | | Unusual quaternary structure of a homodimeric synergistic toxin from mamba snake venom | | Descriptor: | SULFATE ION, Synergistic-type venom protein S2C4 | | Authors: | Jobichen, C, Narumi, A, Sivaraman, J, Kini, R.M. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unusual quaternary structure of a homodimeric synergistic-type toxin from mamba snake venom defines its molecular evolution.
Biochem.J., 477, 2020
|
|
7CBK
 
 | | Structure of Human Neutrophil Elastase Ecotin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ecotin, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Inhibition Mechanism of Ecotin against Neutrophil Elastase by Targeting the Active Site and Secondary Binding Site.
Biochemistry, 59, 2020
|
|
