3WXX
 
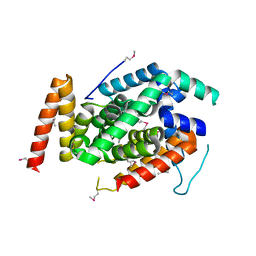 | | Crystal Structure of a T3SS complex from Aeromonas hydrophila | | Descriptor: | AcrH, AopB, MAGNESIUM ION | | Authors: | Nguyen, V.S, Jobichen, C, Sivaraman, J, Henry, Y.K.M. | | Deposit date: | 2014-08-12 | | Release date: | 2015-10-14 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of AcrH-AopB Chaperone-Translocator Complex Reveals a Role for Membrane Hairpins in Type III Secretion System Translocon Assembly
Structure, 23, 2015
|
|
1KAR
 
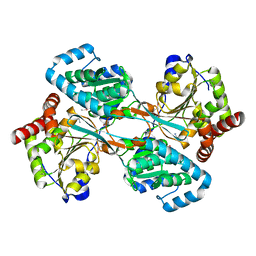 | | L-HISTIDINOL DEHYDROGENASE (HISD) STRUCTURE COMPLEXED WITH HISTAMINE (INHIBITOR), ZINC AND NAD (COFACTOR) | | Descriptor: | HISTAMINE, Histidinol dehydrogenase, ZINC ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J.D, Cygler, M. | | Deposit date: | 2001-11-02 | | Release date: | 2002-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KAE
 
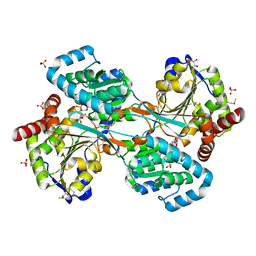 | | L-HISTIDINOL DEHYDROGENASE (HISD) STRUCTURE COMPLEXED WITH L-HISTIDINOL (SUBSTRATE), ZINC AND NAD (COFACTOR) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Histidinol dehydrogenase, ... | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J.D, Cygler, M. | | Deposit date: | 2001-11-01 | | Release date: | 2002-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KAH
 
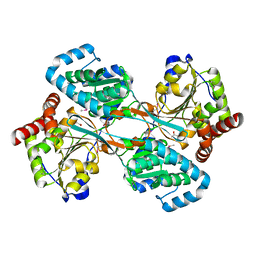 | | L-HISTIDINOL DEHYDROGENASE (HISD) STRUCTURE COMPLEXED WITH L-HISTIDINE (PRODUCT), ZN AND NAD (COFACTOR) | | Descriptor: | HISTIDINE, Histidinol dehydrogenase, ZINC ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J.D, Cygler, M. | | Deposit date: | 2001-11-02 | | Release date: | 2002-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LKZ
 
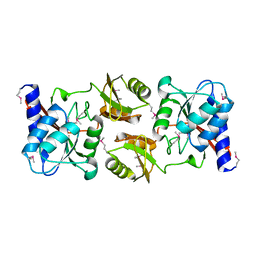 | | Crystal structure of D-ribose-5-phosphate isomerase (RpiA) from Escherichia coli. | | Descriptor: | Ribose 5-phosphate isomerase A | | Authors: | Rangarajan, E.S, Sivaraman, J, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-04-26 | | Release date: | 2002-05-08 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of D-ribose-5-phosphate isomerase (RpiA) from Escherichia coli
Proteins, 48, 2002
|
|
1NQC
 
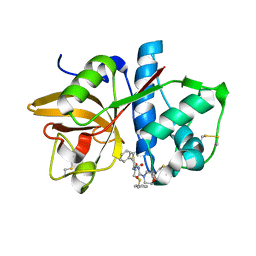 | | Crystal structures of Cathepsin S inhibitor complexes | | Descriptor: | Cathepsin S, N-[(1R)-2-(BENZYLSULFANYL)-1-FORMYLETHYL]-N-(MORPHOLIN-4-YLCARBONYL)-L-PHENYLALANINAMIDE | | Authors: | Pauly, T.A, Sulea, T, Ammirati, M, Sivaraman, J, Danley, D.E, Griffor, M.C, Kamath, A.V, Wang, I.K, Laird, E.R, Menard, R, Cygler, M, Rath, V.L. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specificity determinants of human cathepsin s revealed
by crystal structures of complexes.
Biochemistry, 42, 2003
|
|
3BJW
 
 | | Crystal Structure of ecarpholin S complexed with suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Phospholipase A2 | | Authors: | Zhou, X, Sivaraman, J. | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Myotoxic Ecarpholin S from Echis carinatus Venom
Biophys.J., 95, 2008
|
|
3E3C
 
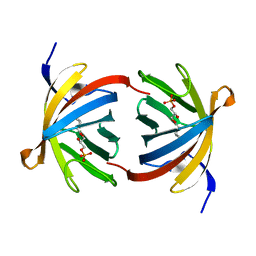 | | Structure of GrlR-lipid complex | | Descriptor: | (2R)-2-HYDROXY-3-(PHOSPHONOOXY)PROPYL HEXANOATE, L0044 | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2008-08-07 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification and characterization of the lipid binding property of GrlR, a locus of enterocyte effacement regulator.
Biochem.J., 2009
|
|
3EBK
 
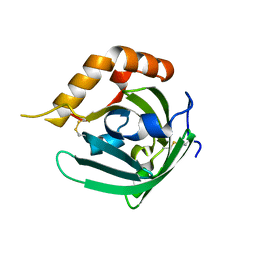 | | Crystal structure of major allergens, Bla g 4 from cockroaches | | Descriptor: | Allergen Bla g 4 | | Authors: | Tan, Y.W, Chan, S.L, Chew, F.T, Sivaraman, J, Mok, Y.K. | | Deposit date: | 2008-08-28 | | Release date: | 2008-12-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of two major allergens, Bla g 4 and Per a 4, from cockroaches and their IgE binding epitopes.
J.Biol.Chem., 284, 2008
|
|
3EBW
 
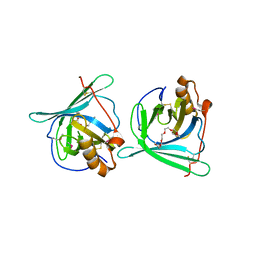 | | Crystal structure of major allergens, Per a 4 from cockroaches | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Per a 4 allergen | | Authors: | Tan, Y.W, Chan, S.L, Chew, F.T, Sivaraman, J, Mok, Y.K. | | Deposit date: | 2008-08-28 | | Release date: | 2008-12-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of two major allergens, Bla g 4 and Per a 4, from cockroaches and their IgE binding epitopes.
J.Biol.Chem., 284, 2008
|
|
3ELP
 
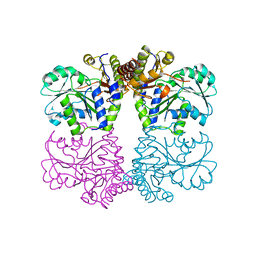 | | Structure of cystationine gamma lyase | | Descriptor: | Cystathionine gamma-lyase | | Authors: | Sun, Q, Sivaraman, J. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the inhibition mechanism of human cystathionine gamma-lyase, an enzyme responsible for the production of H(2)S
J.Biol.Chem., 284, 2009
|
|
3H8B
 
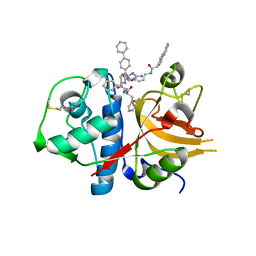 | | A combined crystallographic and molecular dynamics study of cathepsin-L retro-binding inhibitors(compound 9) | | Descriptor: | Cathepsin L1, N~2~,N~6~-bis(biphenyl-4-ylacetyl)-L-lysyl-D-arginyl-N-(2-phenylethyl)-L-phenylalaninamide | | Authors: | Tulsidas, S.R, Chowdhury, S.F, Kumar, S, Joseph, L, Purisima, E.O, Sivaraman, J. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A combined crystallographic and molecular dynamics study of cathepsin L retrobinding inhibitors
J.Med.Chem., 52, 2009
|
|
3H89
 
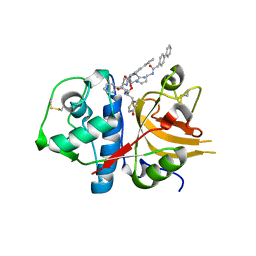 | | A combined crystallographic and molecular dynamics study of cathepsin-L retro-binding inhibitors(compound 4) | | Descriptor: | Cathepsin L1, N~2~,N~6~-bis(biphenyl-4-ylacetyl)-L-lysyl-D-arginyl-N-(2-phenylethyl)-L-tyrosinamide | | Authors: | Tulsidas, S.R, Chowdhury, S.F, Kumar, S, Joseph, L, Purisima, E.O, Sivaraman, J. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A combined crystallographic and molecular dynamics study of cathepsin L retrobinding inhibitors
J.Med.Chem., 52, 2009
|
|
3O4F
 
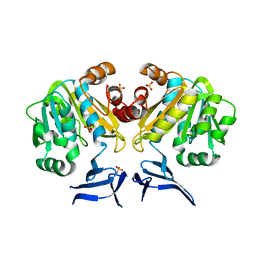 | | Crystal Structure of Spermidine Synthase from E. coli | | Descriptor: | SULFATE ION, Spermidine synthase | | Authors: | Zhou, X, Tkaczuk, K.L, Chruszcz, M, Chua, T.K, Minor, W, Sivaraman, J. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of Escherichia coli spermidine synthase SpeE reveals a unique substrate-binding pocket
J.Struct.Biol., 169, 2010
|
|
4N7C
 
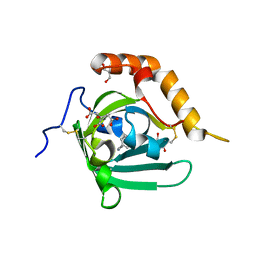 | | Structural re-examination of native Bla g 4 | | Descriptor: | 4-(2-aminoethyl)phenol, Bla g 4 allergen variant 1, CITRIC ACID, ... | | Authors: | Offermann, L.R, Chan, S.L, Osinski, T, Tan, Y.W, Chew, F.T, Sivaraman, J, Mok, Y.K, Minor, W, Chruszcz, M. | | Deposit date: | 2013-10-15 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The major cockroach allergen Bla g 4 binds tyramine and octopamine.
Mol.Immunol., 60, 2014
|
|
4N7D
 
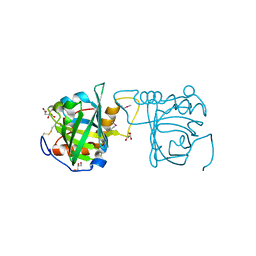 | | Selenomethionine incorporated Bla g 4 | | Descriptor: | Bla g 4 allergen variant 1, CITRIC ACID, GLYCEROL | | Authors: | Offermann, L.R, Chan, S.L, Osinski, T, Tan, Y.W, Chew, F.T, Sivaraman, J, Mok, Y.K, Minor, W, Chruszcz, M. | | Deposit date: | 2013-10-15 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The major cockroach allergen Bla g 4 binds tyramine and octopamine.
Mol.Immunol., 60, 2014
|
|
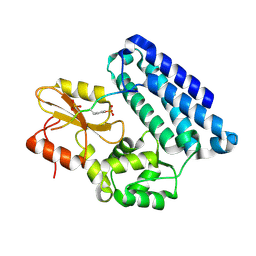
3OB1
 
 | |
3OB2
 
 | |
3OF8
 
 | |
3OF9
 
 | |
3PH0
 
 | | Crystal structure of the heteromolecular chaperone, AscE-AscG, from the type III secretion system in Aeromonas hydrophila | | Descriptor: | AscE, AscG | | Authors: | Chatterjee, C, Kumar, S, Chakraborty, S, Tan, Y.W, Leung, K.Y, Sivaraman, J, Mok, Y.K. | | Deposit date: | 2010-11-03 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the heteromolecular chaperone, AscE-AscG, from the type III secretion system in Aeromonas hydrophila
Plos One, 6, 2011
|
|
3PIS
 
 | |
3PLC
 
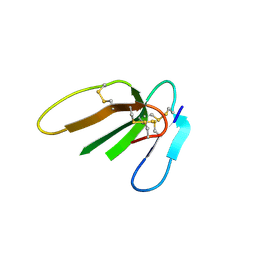 | | Crystal structure of Beta-Cardiotoxin, a novel three-finger cardiotoxin from the venom of Ophiophagus hannah | | Descriptor: | Beta-cardiotoxin OH-27 | | Authors: | Roy, A, Qingxiang, S, Kini, R.M, Sivaraman, J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Identification of a alpha-helical molten globule intermediate and structural characterization of beta-cardiotoxin, an all beta-sheet protein isolated from the venom of Ophiophagus hannah (king cobra).
Protein Sci., 28, 2019
|
|
3PLF
 
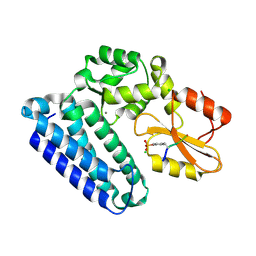 | | Reverse Binding Mode of MetRD peptide complexed with c-Cbl TKB domain | | Descriptor: | CALCIUM ION, CHLORIDE ION, E3 ubiquitin-protein ligase CBL, ... | | Authors: | Sun, Q, Sivaraman, J. | | Deposit date: | 2010-11-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | An adjacent arginine, and the phosphorylated tyrosine in the c-Met receptor target sequence, dictates the orientation of c-Cbl binding
Febs Lett., 585, 2011
|
|
4O7D
 
 | |
