7OVU
 
 | |
7OVV
 
 | | Crystal structure of the Arabidopsis thaliana thialysine acetyltransferase AtNATA2 | | Descriptor: | Probable acetyltransferase NATA1-like, [[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyldisulfanyl]ethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Layer, D, Kopp, J, Sinning, I. | | Deposit date: | 2021-06-15 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the Arabidopsis thaliana thialysine acetyltransferase AtNATA2
To Be Published
|
|
4ADT
 
 | | Crystal structure of plasmodial PLP synthase | | Descriptor: | PHOSPHATE ION, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, PUTATIVE | | Authors: | Guedez, G, Sinning, I, Tews, I. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
7OLD
 
 | | Thermophilic eukaryotic 80S ribosome at pe/E (TI)-POST state | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2021-05-19 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution structures of a thermophilic eukaryotic 80S ribosome reveal atomistic details of translocation.
Nat Commun, 13, 2022
|
|
7OLC
 
 | | Thermophilic eukaryotic 80S ribosome at idle POST state | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2021-05-19 | | Release date: | 2022-01-26 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution structures of a thermophilic eukaryotic 80S ribosome reveal atomistic details of translocation.
Nat Commun, 13, 2022
|
|
4ADS
 
 | | Crystal structure of plasmodial PLP synthase complex | | Descriptor: | PDX2 PROTEIN, PHOSPHATE ION, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, ... | | Authors: | Guedez, G, Sinning, I, Tews, I. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
4ADU
 
 | | Crystal structure of plasmodial PLP synthase with bound R5P intermediate | | Descriptor: | 1,2-ETHANEDIOL, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, PUTATIVE, ... | | Authors: | Guedez, G, Sinning, I, Tews, I. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
4ZN4
 
 | |
2YHS
 
 | | Structure of the E. coli SRP receptor FtsY | | Descriptor: | 1,2-ETHANEDIOL, CELL DIVISION PROTEIN FTSY | | Authors: | Stjepanovic, G, Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2011-05-05 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Lipids Trigger a Conformational Switch that Regulates Signal Recognition Particle (Srp)-Mediated Protein Targeting.
J.Biol.Chem., 286, 2011
|
|
5AFF
 
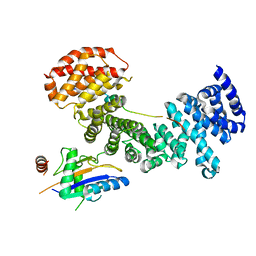 | | Symportin 1 chaperones 5S RNP assembly during ribosome biogenesis by occupying an essential rRNA binding site | | Descriptor: | RIBOSOMAL PROTEIN L11, RIBOSOMAL PROTEIN L5, SYMPORTIN 1 | | Authors: | Calvino, F.R, Kharde, S, Wild, K, Bange, G, Sinning, I. | | Deposit date: | 2015-01-21 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | Symportin 1 Chaperones 5S Rnp Assembly During Ribosome Biogenesis by Occupying an Essential Rrna-Binding Site.
Nat.Commun., 6, 2015
|
|
2FH5
 
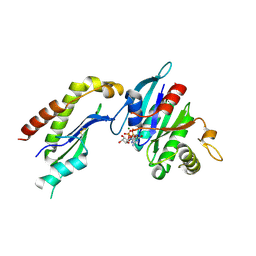 | | The Structure of the Mammalian SRP Receptor | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Signal recognition particle receptor alpha subunit, ... | | Authors: | Schlenker, O, Wild, K, Sinning, I. | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of the mammalian signal recognition particle (SRP) receptor as prototype for the interaction of small GTPases with Longin domains.
J.Biol.Chem., 281, 2006
|
|
6Y31
 
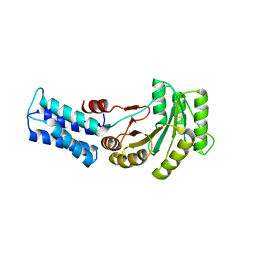 | | NG domain of human SRP54 T117 deletion mutant | | Descriptor: | Signal recognition particle 54 kDa protein | | Authors: | Juaire, K.D, Lapouge, K, Becker, M.M.M, Kotova, I, Haas, M, Carapito, R, Wild, K, Bahram, S, Sinning, I. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | Structural and Functional Impact of SRP54 Mutations Causing Severe Congenital Neutropenia.
Structure, 29, 2021
|
|
4P3G
 
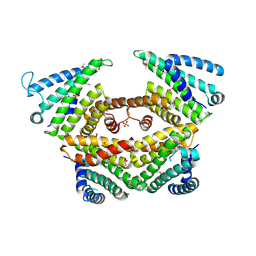 | |
4P3F
 
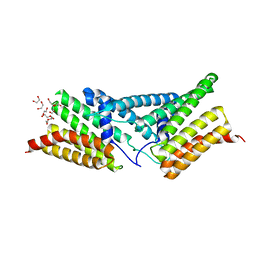 | | Structure of the human SRP68-RBD | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Signal recognition particle subunit SRP68 | | Authors: | Grotwinkel, J.T, Wild, K, Sinning, I. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | SRP RNA remodeling by SRP68 explains its role in protein translocation.
Science, 344, 2014
|
|
4P3E
 
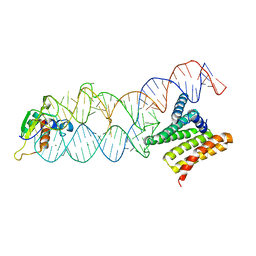 | | Structure of the human SRP S domain | | Descriptor: | MAGNESIUM ION, SRP RNA (124-mer), Signal recognition particle 19 kDa protein, ... | | Authors: | Grotwinkel, J.T, Wild, K, Sinning, I. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | SRP RNA remodeling by SRP68 explains its role in protein translocation.
Science, 344, 2014
|
|
6Y32
 
 | | Structure of the GTPase heterodimer of human SRP54 and SRalpha | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, SULFATE ION, ... | | Authors: | Juaire, K.D, Becker, M.M.M, Wild, K, Sinning, I. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Functional Impact of SRP54 Mutations Causing Severe Congenital Neutropenia.
Structure, 29, 2021
|
|
6Y2Z
 
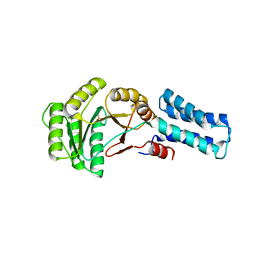 | | NG domain of human SRP54 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Signal recognition particle 54 kDa protein | | Authors: | Juaire, K.D, Wild, K, Sinning, I. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Impact of SRP54 Mutations Causing Severe Congenital Neutropenia.
Structure, 29, 2021
|
|
6Y30
 
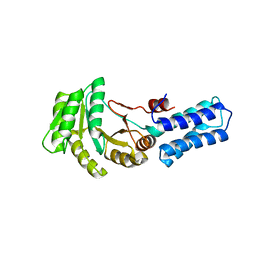 | | NG domain of human SRP54 T115A mutant | | Descriptor: | Signal recognition particle 54 kDa protein | | Authors: | Juaire, K.D, Wild, K, Sinning, I. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Functional Impact of SRP54 Mutations Causing Severe Congenital Neutropenia.
Structure, 29, 2021
|
|
5LNS
 
 | | Crystal structure of Arabidopsis thaliana Pdx1-R5P complex | | Descriptor: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, RIBULOSE-5-PHOSPHATE | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5LNU
 
 | | Crystal structure of Arabidopsis thaliana Pdx1-I320 complex | | Descriptor: | (4~{S})-4-azanyl-5-oxidanyl-pent-1-en-3-one, PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5LNR
 
 | | Crystal structure of Arabidopsis thaliana Pdx1-PLP complex | | Descriptor: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal 5'-phosphate synthase subunit PDX1.3 | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2017-02-22 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5LNW
 
 | | Crystal structure of Arabidopsis thaliana Pdx1-I320-G3P complex | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, GLYCEROL, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
3SJB
 
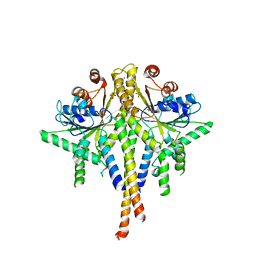 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3SJC
 
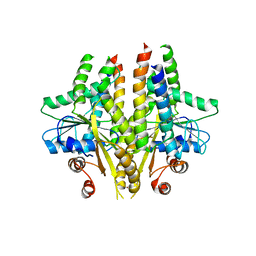 | |
3SJA
 
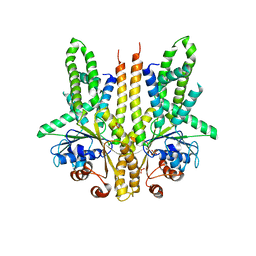 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
