5CIX
 
 | | Structure of the complex of type 1 Ribosome inactivating protein with triethanolamine at 1.88 Angstrom resolution | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Singh, P.K, Pandey, S, Tyagi, T.K, Singh, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-13 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure of the complex of type 1 Ribosome inactivating protein with triethanolamine at 1.88 Angstrom resolution.
To Be Published
|
|
5Y48
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with Pyrimidine-2,4-dione at 1.70 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, URACIL | | Authors: | Singh, P.K, Pandey, S, Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
7Y3U
 
 | | Crystal structure of the complex of Lactoperoxidase with Nitric oxide at 2.50A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Singh, P.K, Viswanathan, V, Ahmad, N, Rani, C, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-06-13 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of Lactoperoxidase with Nitric oxide at 2.50A resolution
To Be Published
|
|
6L9T
 
 | | Crystal structure of the complex of bovine lactoperoxidase with OSCN at 1.89 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Viswanathan, V, Pandey, N, Singh, A, Sinha, M, Singh, R.P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the complex of bovine lactoperoxidase with OSCN at 1.89 A resolution
To Be Published
|
|
5DWF
 
 | | Crystal structure of the complex of Peptidoglycan recognition protein, PGRP-S from camel with ethylene glycol at 1.83 A resolution | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Singh, P.K, Yadav, S.P, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-09-22 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the complex of Peptidoglycan recognition protein, PGRP-S from camel with ethylene glycol at 1.83 A resolution
To Be Published
|
|
6L5G
 
 | | Crystal structure of yak lactoperoxidase with disordered heme moiety at 2.50 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, P.K, Rani, C, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Potassium-induced partial inhibition of lactoperoxidase: structure of the complex of lactoperoxidase with potassium ion at 2.20 angstrom resolution.
J.Biol.Inorg.Chem., 26, 2021
|
|
6KY7
 
 | | Crystal structure of yak lactoperoxidase at 2.27 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, P.K, Viswanathan, V, Sharma, P, Rani, C, Sharma, S, Singh, T.P. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of yak lactoperoxidase at 2.27 A resolution
To Be Published
|
|
7C73
 
 | | Crystal structure of yak lactoperoxidase using data obtained from crystals soaked in MgCl2 at 2.70 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lactoperoxidase, ... | | Authors: | Singh, P.K, Pandey, S.N, Rani, C, Ahmad, N, Viswanathan, V, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Potassium-induced partial inhibition of lactoperoxidase: structure of the complex of lactoperoxidase with potassium ion at 2.20 angstrom resolution.
J.Biol.Inorg.Chem., 26, 2021
|
|
7C75
 
 | | Crystal structure of yak lactoperoxidase with partially coordinated Na ion in the distal heme cavity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lactoperoxidase, ... | | Authors: | Singh, P.K, Viswanathan, V, Rani, C, Ahmad, N, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Potassium-induced partial inhibition of lactoperoxidase: structure of the complex of lactoperoxidase with potassium ion at 2.20 angstrom resolution.
J.Biol.Inorg.Chem., 26, 2021
|
|
7C74
 
 | | Crystal structure of yak lactoperoxidase using data obtained from crystals soaked in CaCl2 at 2.73 A resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, Lactoperoxidase, ... | | Authors: | Singh, P.K, Viswanathan, V, Pandey, S.N, Ahmad, N, Rani, C, Sharma, P, Sharma, P, Singh, T.P. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Potassium-induced partial inhibition of lactoperoxidase: structure of the complex of lactoperoxidase with potassium ion at 2.20 angstrom resolution.
J.Biol.Inorg.Chem., 26, 2021
|
|
5B72
 
 | | Crystal structure of bovine lactoperoxidase with a broken covalent bond between Glu258 and heme moiety at 1.98 A resolution. | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sirohi, H.V, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of bovine lactoperoxidase with a partially linked heme moiety at 1.98 angstrom resolution
Biochim. Biophys. Acta, 1865, 2016
|
|
5WV3
 
 | | Crystal structure of bovine lactoperoxidase with a partial Glu258-heme linkage at 2.07 A resolution. | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sirohi, H.V, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis of activation of mammalian heme peroxidases
Prog. Biophys. Mol. Biol., 133, 2018
|
|
8FTE
 
 | |
8FTF
 
 | |
7VE3
 
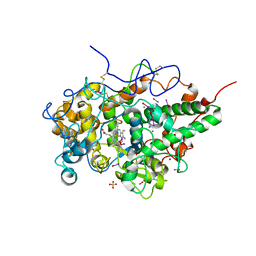 | | Structure of the complex of sheep lactoperoxidase with hypoiodite at 2.70 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Singh, P.K, Yamini, S, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence of the oxidation of iodide ion into hyper-reactive hypoiodite ion by mammalian heme lactoperoxidase.
Protein Sci., 31, 2022
|
|
8VKR
 
 | | CW Flagellar Switch Complex with extra density - FliF, FliG, FliM, and FliN forming the C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
8VKQ
 
 | | CW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming the C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
8VIB
 
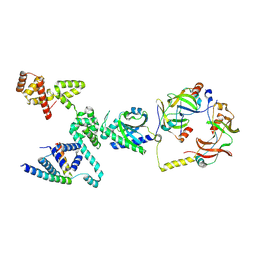 | | CW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming single subunit of the C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2024-01-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
8VID
 
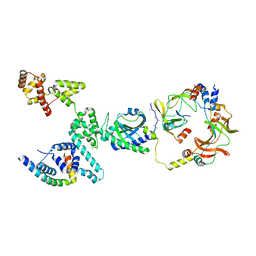 | | CW Flagellar Switch Complex with extra density - FliF, FliG, FliM, and FliN forming single subunit of the C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2024-01-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
5ZGS
 
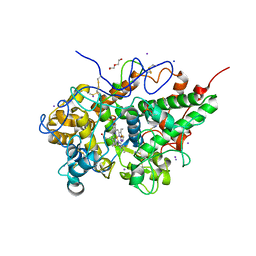 | | Crystal structure of the complex of bovine lactoperoxidase with multiple SCN and OSCN ions in the distal heme cavity | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Singh, R.P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-03-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the complex of bovine lactoperoxidase with multiple SCN and OSCN ions in the distal heme cavity
To Be Published
|
|
5GH0
 
 | | Crystal structure of the complex of bovine lactoperoxidase with mercaptoimidazole at 2.3 A resolution | | Descriptor: | 1,3-dihydroimidazole-2-thione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sirohi, H.V, Singh, A.K, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-06-17 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of anti-thyroid drugs: Binding studies and structure determination of the complex of lactoperoxidase with 2-mercaptoimidazole at 2.30 angstrom resolution
Proteins, 85, 2017
|
|
8T8P
 
 | | 33-mer FliF MS-ring from Salmonella | | Descriptor: | Flagellar M-ring protein | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2023-06-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
8T8O
 
 | | CCW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming 34-mer C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliN, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2023-06-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
5ILX
 
 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, ... | | Authors: | Singh, P.K, Singh, A, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution
To Be Published
|
|
5DUI
 
 | |
