7ENU
 
 | | Crystal structure of iron-saturated C-terminal half of lactoferrin produced proteolytically using pepsin at 2.32A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Singh, J, Maurya, A, Viswanathan, V, Singh, P.K, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.322 Å) | | Cite: | Crystal structure of iron-saturated C-terminal half of lactoferrin produced proteolytically using pepsin at 2.32A resolution
To Be Published
|
|
7EV0
 
 | | Crystal structure of pepsin cleaved C-terminal half of lactoferrin at 2.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Singh, J, Ahmad, M.I, Maurya, A, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pepsin cleaved C-terminal half of lactoferrin at 2.7A resolution
To Be Published
|
|
7VQF
 
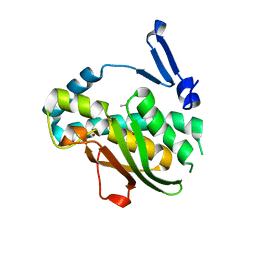 | | Phenol binding protein, MopR | | Descriptor: | ACETATE ION, PHENOL, Phenol sensing regulator, ... | | Authors: | Singh, J, Ray, S, Anand, R. | | Deposit date: | 2021-10-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phenol sensing in nature is modulated via a conformational switch governed by dynamic allostery.
J.Biol.Chem., 298, 2022
|
|
5GJL
 
 | | Solution structure of SUMO from Plasmodium falciparum | | Descriptor: | Uncharacterized protein | | Authors: | Singh, J.S, Shukla, V.K, Gujrati, M, Mishra, R.K, Kumar, A. | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interaction study of SUMO from Plasmodium falciparum
To Be Published
|
|
7F5N
 
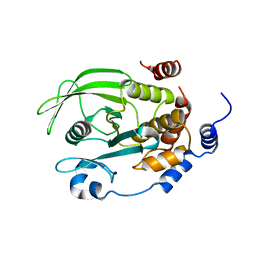 | | Crystal structure of TCPTP catalytic domain | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Singh, J.P, Lin, M.-J, Hsu, S.-F, Lee, C.-C, Meng, T.-C. | | Deposit date: | 2021-06-22 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of TCPTP Unravels an Allosteric Regulatory Role of Helix alpha 7 in Phosphatase Activity.
Biochemistry, 60, 2021
|
|
7F5O
 
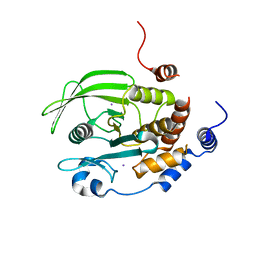 | | Crystal structure of PTPN2 catalytic domain | | Descriptor: | IODIDE ION, Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Singh, J.P, Lin, M.-J, Hsu, S.-F, Lee, C.-C, Meng, T.-C. | | Deposit date: | 2021-06-22 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of TCPTP Unravels an Allosteric Regulatory Role of Helix alpha 7 in Phosphatase Activity.
Biochemistry, 60, 2021
|
|
7FDW
 
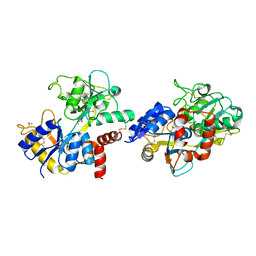 | | Crystal structure of pepsin cleaved lactoferrin C-lobe at 2.28 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Singh, P.K, Singh, J, Maurya, A, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-07-18 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.277 Å) | | Cite: | A Peptide Bond from the Inter-lobe Segment in the Bilobal Lactoferrin Acts as a Preferred Site for Cleavage for Serine Proteases to Generate the Perfect C-lobe: Structure of the Pepsin Hydrolyzed Lactoferrin C-lobe at 2.28 angstrom Resolution.
Protein J., 40, 2021
|
|
7EQU
 
 | | Crystal structure of the C-lobe of lactoferrin produced by limited proteolysis using pepsin at 2.74A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, FE (III) ION, ... | | Authors: | Maurya, A, Singh, J, Sharma, A, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-05-04 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Crystal structure of the C-lobe of lactoferrin produced by limited proteolysis using pepsin at 2.74A resolution
To Be Published
|
|
7EVQ
 
 | | Crystal structure of C-terminal half of lactoferrin obtained by limited proteolysis using pepsin at 2.6 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, ... | | Authors: | Viswanathan, V, Singh, J, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-05-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of C-terminal half of lactoferrin obtained by limited proteolysis using pepsin at 2.6 A resolution
To Be Published
|
|
7C3T
 
 | | Crystal structure of NE0047 (N66Q) mutant in complex with 8-azaguanine | | Descriptor: | 5-AMINO-1H-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-7-OL, Cytidine and deoxycytidylate deaminase zinc-binding region, ZINC ION | | Authors: | Gaded, V, Bitra, A, Singh, J, Anand, R. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure guided mutagenesis reveals the substrate determinants of guanine deaminase.
J.Struct.Biol., 213, 2021
|
|
7C3U
 
 | | Crystal structure of NE0047 (N66A) mutant in complex with 8-azaguanine | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-1H-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-7-OL, Cytidine and deoxycytidylate deaminase zinc-binding region, ... | | Authors: | Gaded, V, Bitra, A, Singh, J, Anand, R. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure guided mutagenesis reveals the substrate determinants of guanine deaminase.
J.Struct.Biol., 213, 2021
|
|
7C3S
 
 | | Crystal structure of NE0047 (E143D) mutant in complex with 8-azaguanine | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-1H-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-7-OL, Cytidine and deoxycytidylate deaminase zinc-binding region, ... | | Authors: | Gaded, V, Bitra, A, Singh, J, Anand, R. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure guided mutagenesis reveals the substrate determinants of guanine deaminase.
J.Struct.Biol., 213, 2021
|
|
7V2N
 
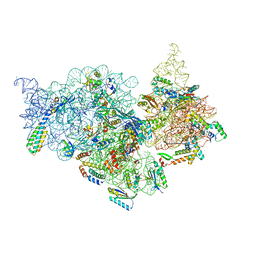 | | T.thermophilus 30S ribosome with KsgA, class K2 | | Descriptor: | 16s ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Raina, R, Singh, J, Anand, R, Vinothkumar, K.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
7V2Q
 
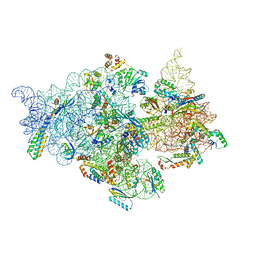 | | T.thermophilus 30S ribosome with KsgA, class K6 | | Descriptor: | 16s ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Raina, R, Singh, J, Anand, R, Vinothkumar, K.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
7V2O
 
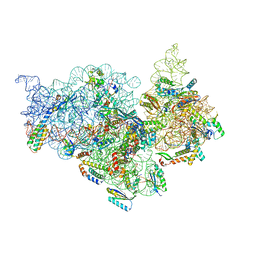 | | T.thermophilus 30S ribosome with KsgA, class K4 | | Descriptor: | 16s ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Raina, R, Singh, J, Anand, R, Vinothkumar, K.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
7V2M
 
 | | T.thermophilus 30S ribosome with KsgA, class K1k4 | | Descriptor: | 16s ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Raina, R, Singh, J, Anand, R, Vinothkumar, K.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
7V2P
 
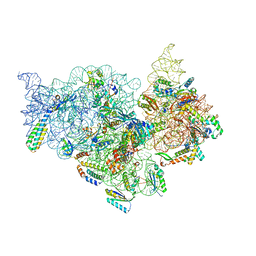 | | T.thermophilus 30S ribosome with KsgA, class K5 | | Descriptor: | 16s ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Raina, R, Singh, J, Anand, R, Vinothkumar, K.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
7V2L
 
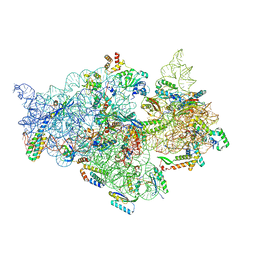 | | T.thermophilus 30S ribosome with KsgA, class K1k2 | | Descriptor: | 16s ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Raina, R, Singh, J, Anand, R, Vinothkumar, K.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
3OD7
 
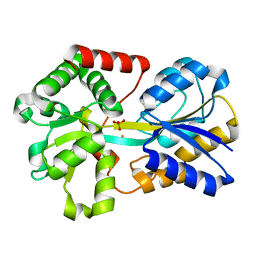 | | Haemophilus influenzae ferric binding protein A -Iron Loaded | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein, PHOSPHATE ION | | Authors: | Khambati, H.K, Moraes, T.F, Singh, J, Shouldice, S.R, Yu, R.H, Schryvers, A.B. | | Deposit date: | 2010-08-11 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The role of vicinal tyrosine residues in the function of Haemophilus influenzae ferric binding protein A.
Biochem.J., 2010
|
|
3ODB
 
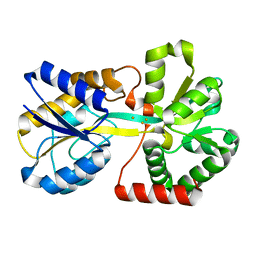 | | Haemophilus influenzae ferric binding protein A -Iron Loaded -open Conformation | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein | | Authors: | Khambati, H.K, Moraes, T.F, Singh, J, Shouldice, S.R, Yu, R.H, Schryvers, A.B. | | Deposit date: | 2010-08-11 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The role of vicinal tyrosine residues in the function of Haemophilus influenzae ferric binding protein A.
Biochem.J., 2010
|
|
3OYP
 
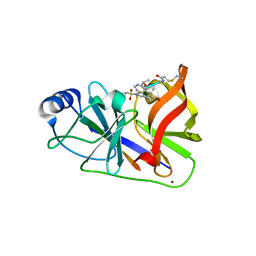 | | HCV NS3/4A in complex with ligand 3 | | Descriptor: | Non-structural protein 4A, Peptidomimetic inhibitor, Serine protease NS3, ... | | Authors: | Hagel, M, Niu, D, St.Martin, T, Sheets, M.P, Qiao, L, Bernard, H, Karp, R.M, Zhu, Z, Labenski, M.T, Chaturvedi, P.C, Nacht, M, Westlin, W.F, Petter, R.C, Singh, J. | | Deposit date: | 2010-09-23 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Selective irreversible inhibition of a protease by targeting a noncatalytic cysteine.
Nat.Chem.Biol., 7, 2011
|
|
3ZIM
 
 | | Discovery of a potent and isoform-selective targeted covalent inhibitor of the lipid kinase PI3Kalpha | | Descriptor: | 1-[4-[[2-(1H-indazol-4-yl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidin-6-yl]methyl]piperazin-1-yl]-6-methyl-hept-5-ene-1,4- dione, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM | | Authors: | Nacht, M, Qiao, L, Sheets, M.P, Martin, T.S, Labenski, M, Mazdiyasni, H, Karp, R, Zhu, Z, Chaturvedi, P, Bhavsar, D, Niu, D, Westlin, W, Petter, R.C, Medikonda, A.P, Jestel, A, Blaesse, M, Singh, J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of a Potent and Isoform-Selective Targeted Covalent Inhibitor of the Lipid Kinase Pi3Kalpha
J.Med.Chem., 56, 2013
|
|
