5FF1
 
 | | Two way mode of binding of antithyroid drug methimazole to mammalian heme peroxidases: Structure of the complex of lactoperoxidase with methimazole at 1.97 Angstrom resolution | | Descriptor: | 1-METHYL-1,3-DIHYDRO-2H-IMIDAZOLE-2-THIONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, R.P, Singh, A, Sirohi, H, Singh, A.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-12-17 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Dual binding mode of antithyroid drug methimazole to mammalian heme peroxidases - structural determination of the lactoperoxidase-methimazole complex at 1.97 angstrom resolution.
Febs Open Bio, 6, 2016
|
|
2HPZ
 
 | | Crystal structure of proteinase K complex with a synthetic peptide KLKLLVVIRLK at 1.69 A resolution | | Descriptor: | 11-mer synthetic peptide, CALCIUM ION, NITRATE ION, ... | | Authors: | Prem kumar, R, Singh, A.K, Somvanshi, R.K, Singh, N, Sharma, S, Kaur, P, Dey, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-07-18 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of proteinase K complex with a synthetic peptide KLKLLVVIRLK at 1.69 A resolution
To be Published
|
|
2IKC
 
 | | Crystal structure of sheep lactoperoxidase at 3.25 A resolution reveals the binding sites for formate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sheikh, I.A, Singh, N, Singh, A.K, Sharma, S, Singh, T.P. | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of sheep lactoperoxidase at 3.25 A resolution reveals the binding sites for formate
To be Published
|
|
2HD4
 
 | | Crystal structure of proteinase K inhibited by a lactoferrin octapeptide Gly-Asp-Glu-Gln-Gly-Glu-Asn-Lys at 2.15 A resolution | | Descriptor: | 8-mer Peptide from Lactotransferrin, ACETIC ACID, CALCIUM ION, ... | | Authors: | Prem Kumar, R, Singh, A.K, Singh, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2006-06-20 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of proteinase K inhibited by a lactoferrin octapeptide Gly-Asp-Glu-Gln-Gly-Glu-Asn-Lys at 2.15 A resolution
To be Published
|
|
2GJM
 
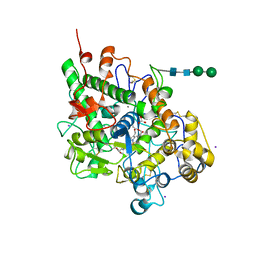 | | Crystal structure of Buffalo lactoperoxidase at 2.75A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBONATE ION, ... | | Authors: | Sheikh, I.A, Ethayathulla, A.S, Singh, A.K, Singh, N, Sharma, S, Singh, T.P. | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of Buffalo lactoperoxidase at 2.75A resolution
To be Published
|
|
7YOE
 
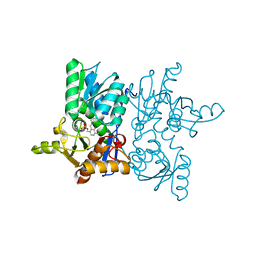 | |
7YOF
 
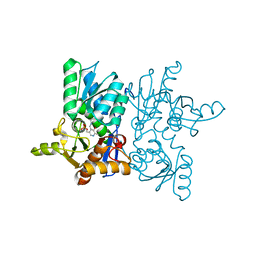 | |
7YOG
 
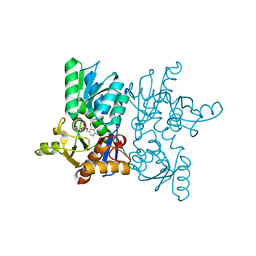 | |
7YOI
 
 | |
7YOK
 
 | | Crystal Structure of Tetra mutant (D67E, A68P, L98I, A301S) tetra mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae at 2.8 A | | Descriptor: | Cysteine synthase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Crystal Structure of Tetra mutant (D67E, A68P, L98I, A301S) tetra mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae at 2.8 A
To Be Published
|
|
7YOL
 
 | | Crystal structure of tetra mutant (D67E, A68P, L98I, A301S) of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide of serine acetyltransferase from Haemophilus influenzae at 2.4 A | | Descriptor: | Cysteine synthase, Peptide from Serine acetyltransferase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of tetra mutant (D67E, A68P, L98I, A301S) of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide from serine acetyltransferase of Haemophilus influenzae at 2.4 A
To Be Published
|
|
7YOD
 
 | |
7YOH
 
 | |
7YOM
 
 | | Crystal structure of tetra mutant (D67E,A68P,L98I,A301S) of O-acetylserine sulfhydrylase from Salmonella typhimurium in complex with high-affinity inhibitory peptide from serine acetyltransferase of Salmonella typhimurium at 2.8 A | | Descriptor: | Cysteine synthase, peptide from serine acetyltransferase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of I88L single mutant of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide from serine acetyltransferase of Salmonella typhimurium at 2.14 A
To Be Published
|
|
5GH0
 
 | | Crystal structure of the complex of bovine lactoperoxidase with mercaptoimidazole at 2.3 A resolution | | Descriptor: | 1,3-dihydroimidazole-2-thione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sirohi, H.V, Singh, A.K, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-06-17 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of anti-thyroid drugs: Binding studies and structure determination of the complex of lactoperoxidase with 2-mercaptoimidazole at 2.30 angstrom resolution
Proteins, 85, 2017
|
|
4UXG
 
 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, R32 native crystal | | Descriptor: | LARGE TAIL FIBER PROTEIN P34 | | Authors: | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
4UXF
 
 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, P21 native crystal | | Descriptor: | GLYCEROL, LARGE TAIL FIBER PROTEIN P34 | | Authors: | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
5IWR
 
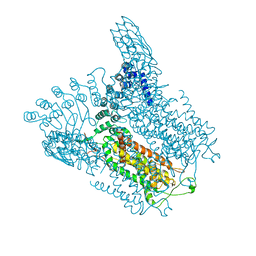 | | Structure of Transient Receptor Potential (TRP) channel TRPV6 in the presence of barium | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, BARIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Saotome, K, Singh, A.K, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2016-03-22 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Crystal structure of the epithelial calcium channel TRPV6.
Nature, 534, 2016
|
|
5IWT
 
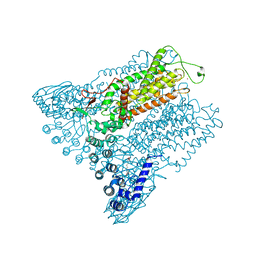 | | Structure of Transient Receptor Potential (TRP) channel TRPV6 in the presence of gadolinium | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, GADOLINIUM ATOM, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Saotome, K, Singh, A.K, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2016-03-22 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of the epithelial calcium channel TRPV6.
Nature, 534, 2016
|
|
5IWK
 
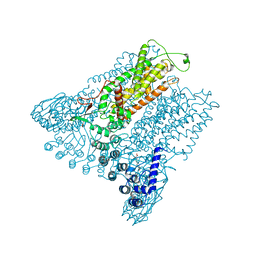 | | Structure of Transient Receptor Potential (TRP) channel TRPV6 | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Saotome, K, Singh, A.K, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2016-03-22 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.247 Å) | | Cite: | Crystal structure of the epithelial calcium channel TRPV6.
Nature, 534, 2016
|
|
5IWP
 
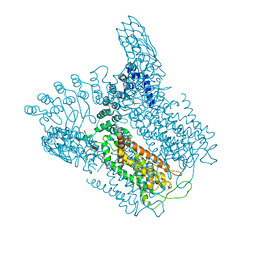 | | Structure of Transient Receptor Potential (TRP) channel TRPV6 in the presence of calcium | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Saotome, K, Singh, A.K, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2016-03-22 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Crystal structure of the epithelial calcium channel TRPV6.
Nature, 534, 2016
|
|
6PW5
 
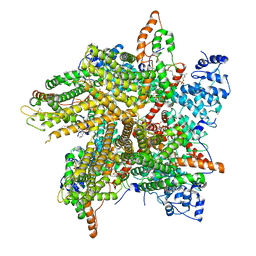 | | Cryo-EM Structure of Thermo-Sensitive TRP Channel TRP1 from the Alga Chlamydomonas reinhardtii in Nanodiscs | | Descriptor: | (2S)-3-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, TRP-like ion channel, ... | | Authors: | McGoldrick, L.L, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-25 | | Last modified: | 2019-12-04 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the thermo-sensitive TRP channel TRP1 from the alga Chlamydomonas reinhardtii.
Nat Commun, 10, 2019
|
|
6PW4
 
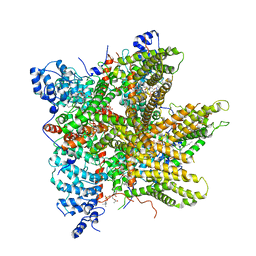 | | Cryo-EM Structure of Thermo-Sensitive TRP Channel TRP1 from the Alga Chlamydomonas reinhardtii in Detergent | | Descriptor: | (2S)-3-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | McGoldrick, L.L, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structure of the thermo-sensitive TRP channel TRP1 from the alga Chlamydomonas reinhardtii.
Nat Commun, 10, 2019
|
|
5BN7
 
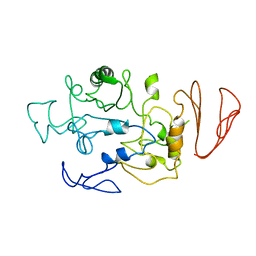 | | Crystal structure of maltodextrin glucosidase from E.coli at 3.7 A resolution | | Descriptor: | Maltodextrin glucosidase | | Authors: | Shukla, P.K, Pastor, A, Singh, A.K, Sharma, S, Singh, T.P, Chaudhuri, T.K. | | Deposit date: | 2015-05-25 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Role of N-terminal region of Escherichia coli maltodextrin glucosidase in folding and function of the protein
Biochim.Biophys.Acta, 1864, 2016
|
|
7OTX
 
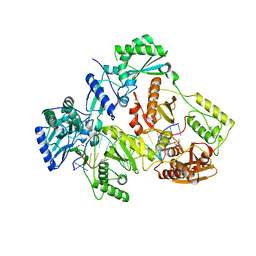 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-257 | | Descriptor: | (S)-2-((3-(6-amino-9H-purin-9-yl)propyl)amino)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
