6PAY
 
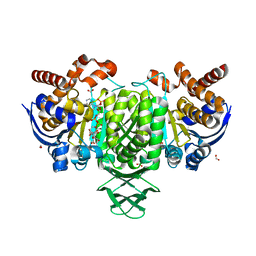 | | Structure of HsICDH1:Mg(II):ICT:NADPH(50%) complex reveals structural basis for observation of half-sites reactivity | | Descriptor: | FORMIC ACID, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Silvaggi, N.R, Melkonian, T.R, Roman, J.V, Moran, G.R. | | Deposit date: | 2019-06-12 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Transient-State Analysis of Human Isocitrate Dehydrogenase I: Accounting for the Interconversion of Active and Non-Active Conformational States.
Biochemistry, 58, 2019
|
|
4ZCD
 
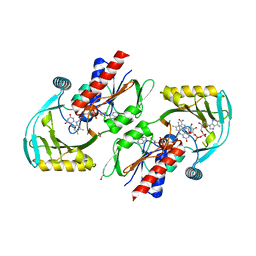 | | Renalase in complex with NAD+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Silvaggi, N.R, Moran, G.R, Roman, J.V. | | Deposit date: | 2015-04-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6605 Å) | | Cite: | Bacterial Renalase: Structure and Kinetics of an Enzyme with 2- and 6-Dihydro-beta-NAD(P) Oxidase Activity from Pseudomonas phaseolicola.
Biochemistry, 54, 2015
|
|
4ZCC
 
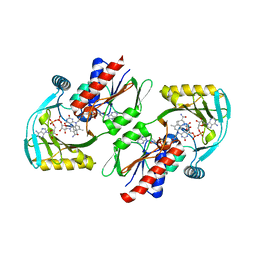 | | Renalase in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Silvaggi, N.R, Moran, G.R, Roman, J.V. | | Deposit date: | 2015-04-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Bacterial Renalase: Structure and Kinetics of an Enzyme with 2- and 6-Dihydro-beta-NAD(P) Oxidase Activity from Pseudomonas phaseolicola.
Biochemistry, 54, 2015
|
|
2FUC
 
 | | Human alpha-Phosphomannomutase 1 with Mg2+ cofactor bound | | Descriptor: | MAGNESIUM ION, Phosphomannomutase 1 | | Authors: | Silvaggi, N.R, Zhang, C, Lu, Z, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2006-01-26 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray crystal structures of human alpha-phosphomannomutase 1 reveal the structural basis of congenital disorder of glycosylation type 1a.
J.Biol.Chem., 281, 2006
|
|
2ILP
 
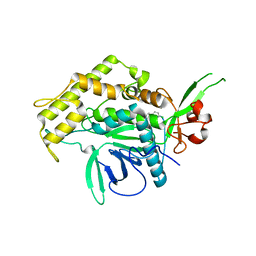 | |
2IMB
 
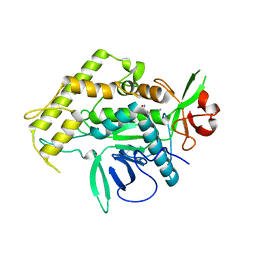 | |
2IMA
 
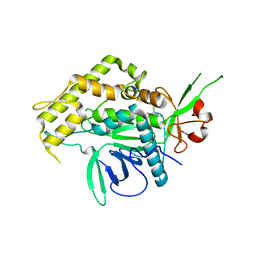 | |
2IMC
 
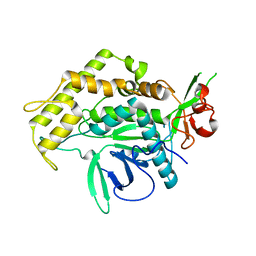 | |
2FUE
 
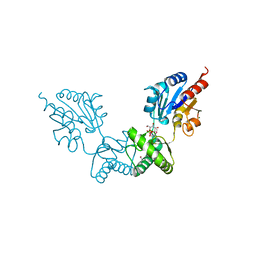 | | Human alpha-Phosphomannomutase 1 with D-mannose 1-phosphate and Mg2+ cofactor bound | | Descriptor: | 1-O-phosphono-alpha-D-mannopyranose, MAGNESIUM ION, Phosphomannomutase 1 | | Authors: | Silvaggi, N.R, Zhang, C, Lu, Z, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2006-01-26 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray crystal structures of human alpha-phosphomannomutase 1 reveal the structural basis of congenital disorder of glycosylation type 1a.
J.Biol.Chem., 281, 2006
|
|
5U8K
 
 | | RitR mutant - C128S | | Descriptor: | Response regulator | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2016-12-14 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | RitR is an archetype for a novel family of redox sensors in the streptococci that has evolved from two-component response regulators and is required for pneumococcal colonization.
PLoS Pathog., 14, 2018
|
|
6EEJ
 
 | | Streptomyces bingchenggensis Aldolase-Dehydratase in covalent complex with dienone product. | | Descriptor: | (3E,5E)-6-(4-nitrophenyl)-2-oxohexa-3,5-dienoic acid, Acetoacetate decarboxylase, FORMIC ACID, ... | | Authors: | Mydy, L.S, Silvaggi, N.R. | | Deposit date: | 2018-08-14 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Mechanistic Studies of theStreptomyces bingchenggensisAldolase-Dehydratase: Implications for Substrate and Reaction Specificity in the Acetoacetate Decarboxylase-like Superfamily.
Biochemistry, 58, 2019
|
|
1YQS
 
 | | Inhibition of the R61 DD-Peptidase by N-benzoyl-beta-sultam | | Descriptor: | 2-(BENZOYLAMINO)ETHANESULFONIC ACID, D-alanyl-D-alanine carboxypeptidase, GLYCEROL | | Authors: | Ahmed, N, Cordaro, M, Laws, A.P, Delmarcelle, M, Silvaggi, N.R, Kelly, J.A, Page, M.I. | | Deposit date: | 2005-02-02 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Inactivation of Bacterial dd-Peptidase by beta-Sultams.
Biochemistry, 44, 2005
|
|
3U3H
 
 | | X-Ray Crystallographic Analysis of D-Xylose Isomerase-Catalyzed Isomerization of (R)-Glyceraldehyde | | Descriptor: | (2R)-propane-1,1,2,3-tetrol, (4R)-2-METHYLPENTANE-2,4-DIOL, FORMIC ACID, ... | | Authors: | Allen, K.N, Silvaggi, N.R, Toteva, M.M, Richard, J.P. | | Deposit date: | 2011-10-05 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Binding Energy and Catalysis by d-Xylose Isomerase: Kinetic, Product, and X-ray Crystallographic Analysis of Enzyme-Catalyzed Isomerization of (R)-Glyceraldehyde.
Biochemistry, 50, 2011
|
|
6VW4
 
 | |
6WEY
 
 | |
1IKI
 
 | | COMPLEX OF STREPTOMYCES R61 DD-PEPTIDASE WITH THE PRODUCTS OF A SPECIFIC PEPTIDOGLYCAN SUBSTRATE FRAGMENT | | Descriptor: | D-ALANINE, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, GLYCYL-L-ALPHA-AMINO-EPSILON-PIMELYL-D-ALANINE | | Authors: | Mcdonough, M.A, Anderson, J.W, Silvaggi, N.R, Pratt, R.F, Knox, J.R, Kelly, J.A. | | Deposit date: | 2001-05-03 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins.
J.Mol.Biol., 322, 2002
|
|
1IKG
 
 | | MICHAELIS COMPLEX OF STREPTOMYCES R61 DD-PEPTIDASE WITH A SPECIFIC PEPTIDOGLYCAN SUBSTRATE FRAGMENT | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, GLYCYL-L-ALPHA-AMINO-EPSILON-PIMELYL-D-ALANYL-D-ALANINE | | Authors: | Mcdonough, M.A, Anderson, J.W, Silvaggi, N.R, Pratt, R.F, Knox, J.R, Kelly, J.A. | | Deposit date: | 2001-05-03 | | Release date: | 2002-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins.
J.Mol.Biol., 322, 2002
|
|
1HVB
 
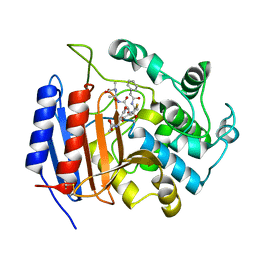 | | CRYSTAL STRUCTURE OF STREPTOMYCES R61 DD-PEPTIDASE COMPLEXED WITH A NOVEL CEPHALOSPORIN ANALOG OF CELL WALL PEPTIDOGLYCAN | | Descriptor: | 5-{3-(S)-(4-(R)-ACETYLAMINO-4-CARBOXY-BUTYRYLAMINO)-3-[1-(R)-(1-(R)-CARBOXY-ETHYLCARBAMOYL)-ETHYLCARBAMOYL]-PROPYL}-2-( CARBOXY-PHENYLACETYLAMINO-METHYL)-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE | | Authors: | McDonough, M.A, Lee, W, Silvaggi, N.R, Mobashery, S, Kelly, J.A. | | Deposit date: | 2001-01-08 | | Release date: | 2001-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A 1.2-A snapshot of the final step of bacterial cell wall biosynthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6XCT
 
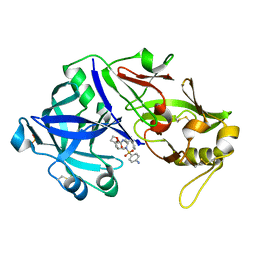 | | Porcine pepsin in complex with amprenavir | | Descriptor: | Pepsin A, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Vuksanovic, N, Silvaggi, N.R. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Porcine pepsin in complex with amprenavir
To Be Published
|
|
6XCZ
 
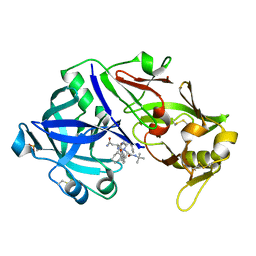 | | Porcine pepsin in complex with saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Pepsin A | | Authors: | Vuksanovic, N, Silvaggi, N.R. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Porcine pepsin in complex with saquinavir
To Be Published
|
|
6XCY
 
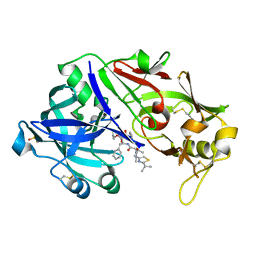 | |
6XD2
 
 | | Porcine pepsin in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Pepsin A | | Authors: | Vuksanovic, N, Silvaggi, N.R. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Porcine pepsin in complex with darunavir
To Be Published
|
|
6C92
 
 | | The structure of MppP soaked with the product 2-ketoarginine | | Descriptor: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
6C9B
 
 | | The structure of MppP soaked with the products 4HKA and 2KA | | Descriptor: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, CHLORIDE ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-26 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
7JQI
 
 | |
