6VTE
 
 | | Naegleria gruberi RNA Ligase K170M mutant with AMP and Mn | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTD
 
 | | Naegleria gruberi RNA ligase R149A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT3
 
 | | Naegleria gruberi RNA ligase K326A mutant apo | | Descriptor: | RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT5
 
 | |
6VTF
 
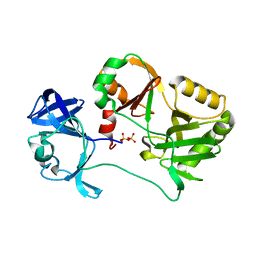 | | Naegleria gruberi RNA ligase with PPi | | Descriptor: | PYROPHOSPHATE 2-, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT9
 
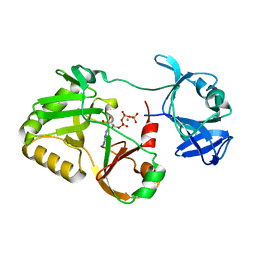 | | Naegleria gruberi RNA ligase E227A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
4YKL
 
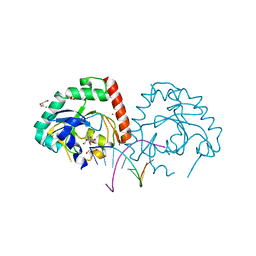 | | Hnt3 in complex with DNA and guanosine | | Descriptor: | Aprataxin-like protein, CHLORIDE ION, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Jacewicz, A, Chauleau, M, Shuman, S. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | DNA3'pp5'G de-capping activity of aprataxin: effect of cap nucleoside analogs and structural basis for guanosine recognition.
Nucleic Acids Res., 43, 2015
|
|
6VT6
 
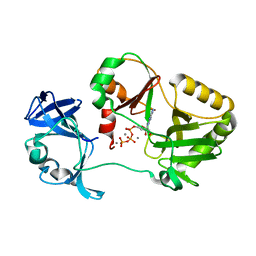 | | Naegleria gruberi RNA ligase K170A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTG
 
 | | Naegleria gruberi RNA ligase E227A mutant apo | | Descriptor: | RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT1
 
 | | Naegleria gruberi RNA ligase D172A mutant apo | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6DTU
 
 | | Maltotetraose bound T. maritima MalE1 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE1 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
5H8A
 
 | | Mmi1 YTH domain | | Descriptor: | YTH domain-containing protein mmi1 | | Authors: | Chatterjee, D, Goldgur, Y, Shuman, S. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Transcription of lncRNA prt, clustered prt RNA sites for Mmi1 binding, and RNA polymerase II CTD phospho-sites govern the repression of pho1 gene expression under phosphate-replete conditions in fission yeast.
Rna, 22, 2016
|
|
6VT0
 
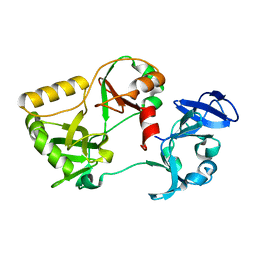 | | Naegleria gruberi RNA ligase K170A mutant apo | | Descriptor: | RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTB
 
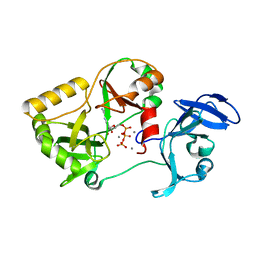 | | Naegleria gruberi RNA ligase K326A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
8TKZ
 
 | |
1VS0
 
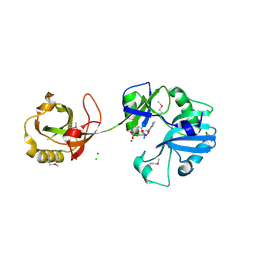 | | Crystal Structure of the Ligase Domain from M. tuberculosis LigD at 2.4A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative DNA ligase-like protein Rv0938/MT0965, ... | | Authors: | Akey, D, Martins, A, Aniukwu, J, Glickman, M.S, Shuman, S, Berger, J.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-01-27 | | Release date: | 2006-02-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Nonhomologous End-joining Function of the Ligase Component of Mycobacterium DNA Ligase D.
J.Biol.Chem., 281, 2006
|
|
6DTQ
 
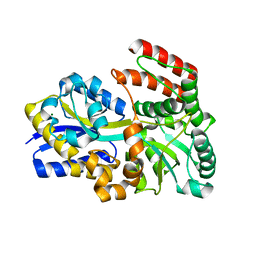 | | Maltose bound T. maritima MalE3 | | Descriptor: | MAGNESIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE3 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6VT8
 
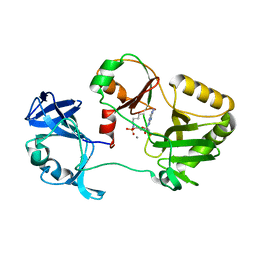 | | Naegleria gruberi RNA ligase E312A mutant with AMP and Mn | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6DTR
 
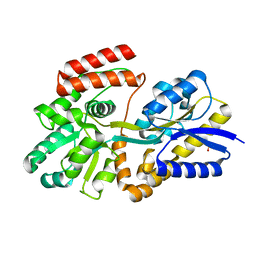 | | Apo T. maritima MalE3 | | Descriptor: | SULFATE ION, maltose-binding protein MalE3 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6E33
 
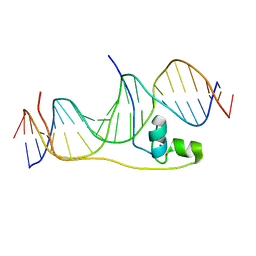 | | Crystal Structure of Pho7-DNA complex | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*GP*AP*AP*TP*GP*TP*CP*CP*GP*AP*AP*GP*GP*AP*T)-3'), DNA (5'-D(*TP*CP*CP*TP*TP*CP*GP*GP*AP*CP*AP*TP*TP*CP*AP*AP*AP*TP*CP*A)-3'), Uncharacterized transcriptional regulatory protein C27B12.11c, ... | | Authors: | Garg, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-07-13 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Distinctive structural basis for DNA recognition by the fission yeast Zn2Cys6 transcription factor Pho7 and its role in phosphate homeostasis.
Nucleic Acids Res., 46, 2018
|
|
6DTS
 
 | | Maltotetraose bound T. maritima MalE2 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE2 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
9DRZ
 
 | |
9DQS
 
 | |
9D8A
 
 | |
9DO4
 
 | |
