4MDF
 
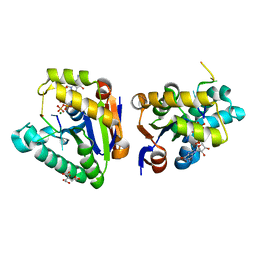 | | Structure of bacterial polynucleotide kinase Michaelis complex bound to GTP and DNA | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*CP*TP*GP*T)-3'), GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shuman, S, Das, U, Wang, L.K, Smith, P, Jacewicz, A. | | Deposit date: | 2013-08-22 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | Structures of bacterial polynucleotide kinase in a Michaelis complex with GTP*Mg2+ and 5'-OH oligonucleotide and a product complex with GDP*Mg2+ and 5'-PO4 oligonucleotide reveal a mechanism of general acid-base catalysis and the determinants of phosphoacceptor recognition.
Nucleic Acids Res., 42, 2014
|
|
4MDE
 
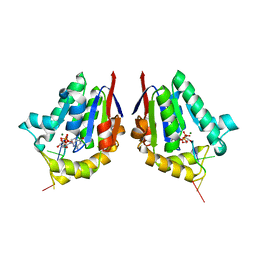 | | Structure of bacterial polynucleotide kinase product complex bound to GDP and DNA | | Descriptor: | DNA (5'-D(P*CP*CP*TP*GP*T)-3'), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shuman, S, Das, U, Wang, L.K, Smith, P, Jacewicz, A. | | Deposit date: | 2013-08-22 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of bacterial polynucleotide kinase in a Michaelis complex with GTP*Mg2+ and 5'-OH oligonucleotide and a product complex with GDP*Mg2+ and 5'-PO4 oligonucleotide reveal a mechanism of general acid-base catalysis and the determinants of phosphoacceptor recognition.
Nucleic Acids Res., 42, 2014
|
|
5TC1
 
 | | In situ structures of the genome and genome-delivery apparatus in ssRNA bacteriophage MS2 | | Descriptor: | Capsid protein, Maturation protein, phage MS2 genome | | Authors: | Dai, X.H, Li, Z.H, Lai, M, Shu, S, Du, Y.S, Zhou, Z.H, Sun, R. | | Deposit date: | 2016-09-13 | | Release date: | 2016-12-07 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus.
Nature, 541, 2017
|
|
7T6D
 
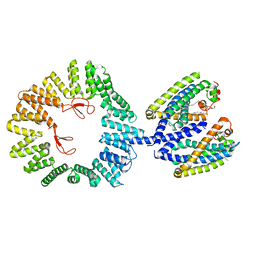 | | CryoEM structure of the YejM/LapB complex | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, Inner membrane protein YejM, ... | | Authors: | Mi, W, Shu, S. | | Deposit date: | 2021-12-13 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Regulatory mechanisms of lipopolysaccharide synthesis in Escherichia coli.
Nat Commun, 13, 2022
|
|
8V24
 
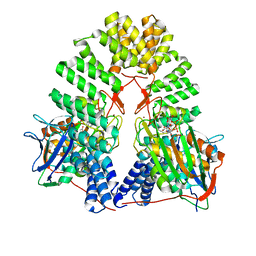 | | LapB cytoplasmic domain in complex with LpxC | | Descriptor: | ACETATE ION, Lipopolysaccharide assembly protein B, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mi, W, Shu, S. | | Deposit date: | 2023-11-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Dual function of LapB (YciM) in regulating Escherichia coli lipopolysaccharide synthesis.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4CKC
 
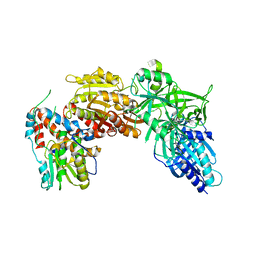 | | Vaccinia virus capping enzyme complexed with SAH (monoclinic form) | | Descriptor: | MRNA-CAPPING ENZYME CATALYTIC SUBUNIT, MRNA-CAPPING ENZYME REGULATORY SUBUNIT, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kyrieleis, O.J.P, Chang, J, de la Pena, M, Shuman, S, Cusack, S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Vaccinia Virus Mrna Capping Enzyme Provides Insights Into the Mechanism and Evolution of the Capping Apparatus.
Structure, 22, 2014
|
|
4W7S
 
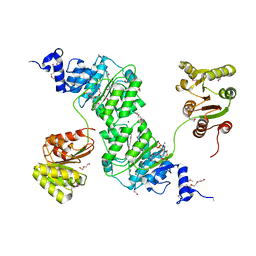 | | Crystal structure of the yeast DEAD-box splicing factor Prp28 at 2.54 Angstroms resolution | | Descriptor: | GLYCEROL, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Jacewicz, A, Smith, P, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Crystal structure, mutational analysis and RNA-dependent ATPase activity of the yeast DEAD-box pre-mRNA splicing factor Prp28.
Nucleic Acids Res., 42, 2014
|
|
1K9F
 
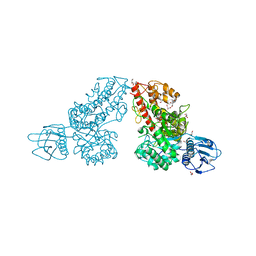 | | Crystal structure of a mutated family-67 alpha-D-glucuronidase (E285N) from Bacillus stearothermophilus T-6, complexed with aldotetraouronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1K9E
 
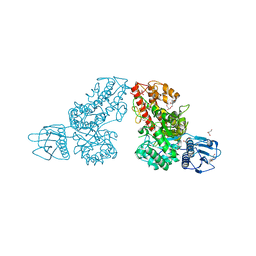 | | Crystal structure of a mutated family-67 alpha-D-glucuronidase (E285N) from Bacillus stearothermophilus T-6, complexed with 4-O-methyl-glucuronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid, GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1K9D
 
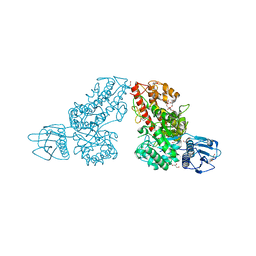 | | The 1.7 A crystal structure of alpha-D-glucuronidase, a family-67 glycoside hydrolase from Bacillus stearothermophilus T-1 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
5COV
 
 | |
9DSY
 
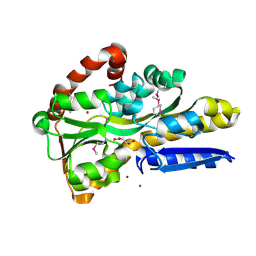 | | Crystal Structure of C4-Dicarboxylate-Binding Periplasmic Protein (PA5167) of Tripartite ATP-independent Periplasmic Transporter Family from Pseudomonas aeruginosa PAO1 in Complex with Succinic Acid | | Descriptor: | C4-dicarboxylate-binding periplasmic protein DctP, SUCCINIC ACID, ZINC ION | | Authors: | Minasov, G, Shukla, S, Shuvalova, L, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-09-30 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of C4-Dicarboxylate-Binding Periplasmic Protein (PA5167) of Tripartite ATP-independent Periplasmic Transporter Family from Pseudomonas aeruginosa PAO1 in Complex with Succinic Acid
To Be Published
|
|
9DTL
 
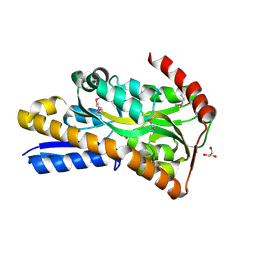 | | Crystal Structure of C4-Dicarboxylate-Binding Protein (PA0884) of Tripartite ATP-independent Periplasmic Transporter Family from Pseudomonas aeruginosa PAO1 in Complex with Succinic Acid | | Descriptor: | C4-dicarboxylate-binding periplasmic protein, GLYCEROL, SUCCINIC ACID | | Authors: | Minasov, G, Shukla, S, Shuvalova, L, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-10-01 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of C4-Dicarboxylate-Binding Protein (PA0884) of Tripartite ATP-independent Periplasmic Transporter Family from Pseudomonas aeruginosa PAO1 in Complex with Succinic Acid
To Be Published
|
|
4WAN
 
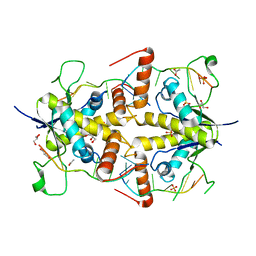 | | Crystal structure of Msl5 protein in complex with RNA at 1.8 A | | Descriptor: | ACETATE ION, Branchpoint-bridging protein, GLYCEROL, ... | | Authors: | Jacewicz, A, Smith, P, Chico, L, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs.
Rna, 21, 2015
|
|
4WAL
 
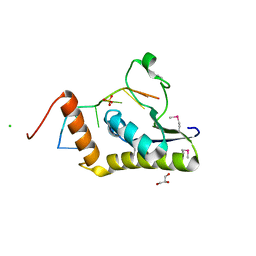 | | Crystal structure of selenomethionine Msl5 protein in complex with RNA at 2.2 A | | Descriptor: | Branchpoint-bridging protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jacewicz, A, Smith, P, Chico, L, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for recognition of intron branchpoint RNA by yeast Msl5 and selective effects of interfacial mutations on splicing of yeast pre-mRNAs.
Rna, 21, 2015
|
|
7KW9
 
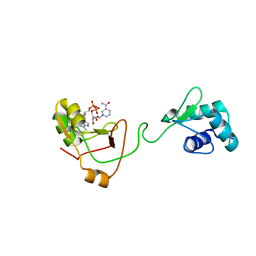 | | NMR Structure of a tRNA 2'-phosphotransferase from Runella slithyformis in complex with NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, tRNA 2'-phosphotransferase | | Authors: | Alphonse, S, Dantuluri, S, Banerjee, A, Shuman, S, Ghose, R. | | Deposit date: | 2020-11-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of Runella slithyformis RNA 2'-phosphotransferase Tpt1 provide insights into NAD+ binding and specificity.
Nucleic Acids Res., 49, 2021
|
|
7KW8
 
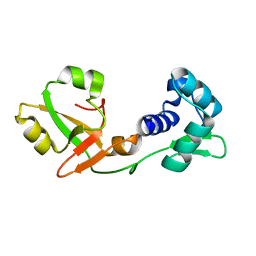 | | NMR Structure of a tRNA 2'-phosphotransferase from Runella slithyformis | | Descriptor: | tRNA 2'-phosphotransferase | | Authors: | Alphonse, S, Dantuluri, S, Banerjee, A, Shuman, S, Ghose, R. | | Deposit date: | 2020-11-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of Runella slithyformis RNA 2'-phosphotransferase Tpt1 provide insights into NAD+ binding and specificity.
Nucleic Acids Res., 49, 2021
|
|
4XBA
 
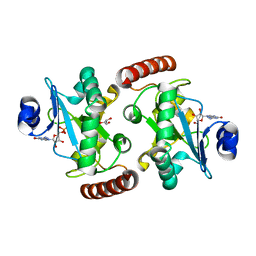 | | Hnt3 | | Descriptor: | Aprataxin-like protein, GLYCEROL, GUANOSINE, ... | | Authors: | Jacewicz, A, Chauleau, M, Shuman, S. | | Deposit date: | 2014-12-16 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | DNA3'pp5'G de-capping activity of aprataxin: effect of cap nucleoside analogs and structural basis for guanosine recognition.
Nucleic Acids Res., 43, 2015
|
|
4CKB
 
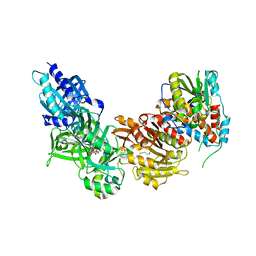 | | Vaccinia virus capping enzyme complexed with GTP and SAH | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MRNA-CAPPING ENZYME CATALYTIC SUBUNIT, MRNA-CAPPING ENZYME REGULATORY SUBUNIT, ... | | Authors: | Kyrieleis, O.J.P, Chang, J, de la Pena, M, Shuman, S, Cusack, S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Vaccinia Virus Mrna Capping Enzyme Provides Insights Into the Mechanism and Evolution of the Capping Apparatus.
Structure, 22, 2014
|
|
4CKE
 
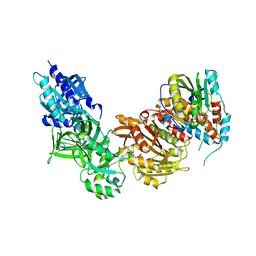 | | Vaccinia virus capping enzyme complexed with SAH in P1 form | | Descriptor: | MRNA-CAPPING ENZYME CATALYTIC SUBUNIT, MRNA-CAPPING ENZYME REGULATORY SUBUNIT, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kyrieleis, O.J.P, Chang, J, de la Pena, M, Shuman, S, Cusack, S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Vaccinia Virus Mrna Capping Enzyme Provides Insights Into the Mechanism and Evolution of the Capping Apparatus.
Structure, 22, 2014
|
|
8DCD
 
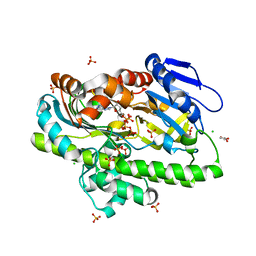 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Zn2+ and GTP | | Descriptor: | CHLORIDE ION, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
8DC9
 
 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Mn2+ and GTP | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
8DCG
 
 | | Structure of guanylylated RNA ligase RtcB from Pyrococcus horikoshii | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
8DCB
 
 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Ni2+ and GTP | | Descriptor: | CHLORIDE ION, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
8DCF
 
 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Cu2+ and GTP | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
