1J27
 
 | | Crystal structure of a hypothetical protein, TT1725, from Thermus thermophilus HB8 at 1.7A resolution | | Descriptor: | hypothetical protein TT1725 | | Authors: | Seto, A, Shirouzu, M, Terada, T, Murayama, K, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-26 | | Release date: | 2003-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a hypothetical protein, TT1725, from Thermus thermophilus HB8 at 1.7 A resolution
Proteins, 53, 2003
|
|
5B6O
 
 | | Crystal structure of MS8104 | | Descriptor: | 3C-like proteinase | | Authors: | Wang, H, Kim, Y, Muramatsu, T, Takemoto, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
3AQV
 
 | | Human AMP-activated protein kinase alpha 2 subunit kinase domain (T172D) complexed with compound C | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine | | Authors: | Handa, N, Takagi, T, Saijo, S, Kishishita, S, Toyama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for compound C inhibition of the human AMP-activated protein kinase alpha 2 subunit kinase domain
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2ZV2
 
 | | Crystal structure of human calcium/calmodulin-dependent protein kinase kinase 2, beta, CaMKK2 kinase domain in complex with STO-609 | | Descriptor: | 7-oxo-7H-benzimidazo[2,1-a]benz[de]isoquinoline-3-carboxylic acid, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Yoshikawa, S, Kukimoto-niino, M, Shirouzu, M, Suzuki, A, Lee, S, Minokoshi, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-10-31 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Ca2+/calmodulin-dependent protein kinase kinase in complex with the inhibitor STO-609
J.Biol.Chem., 286, 2011
|
|
8JDK
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Asp(ManQ34) and mRNA(GAU) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2023-05-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
8JDM
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAU) (rotated state) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2023-05-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
1IUJ
 
 | | The structure of TT1380 protein from thermus thermophilus | | Descriptor: | ZINC ION, hypothetical protein TT1380 | | Authors: | Wada, T, Shirouzu, M, Park, S.-Y, Tame, J.R.H, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-05 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the conserved hypothetical protein TT1380 from Thermus thermophilus HB8
Proteins, 55, 2004
|
|
8JDL
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAU) (non-rotated state) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2023-05-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
8JDJ
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Asp(Q34) and mRNA(GAU) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2023-05-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
5B0U
 
 | |
7DQE
 
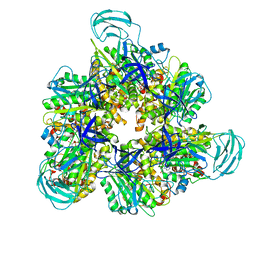 | | Crystal structure of the ADP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Maruyama, S, Nakamoto, K, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | The Combination of High-Speed AFM and X-ray Crystallography Reveals Rotary Catalytic Mechanism of Shaftless V1-ATPase
To Be Published
|
|
7DQD
 
 | | Crystal structure of the AMP-PNP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.383 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalysis
To Be Published
|
|
1IUK
 
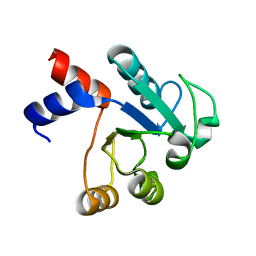 | | The structure of native ID.343 from Thermus thermophilus | | Descriptor: | hypothetical protein TT1466 | | Authors: | Wada, T, Shirouzu, M, Park, S.-Y, Tame, J.R, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-05 | | Release date: | 2003-07-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a conserved CoA-binding protein synthesized by a cell-free system.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1IUG
 
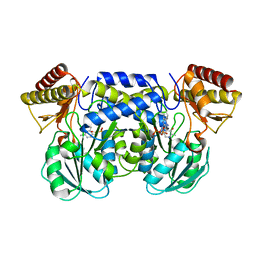 | | The crystal structure of aspartate aminotransferase which belongs to subgroup IV from Thermus thermophilus | | Descriptor: | PHOSPHATE ION, putative aspartate aminotransferase | | Authors: | Katsura, Y, Shirouzu, M, Yamaguchi, H, Ishitani, R, Nureki, O, Kuramitsu, S, Hayashi, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-04 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative aspartate aminotransferase belonging to subgroup IV.
Proteins, 55, 2004
|
|
1IUL
 
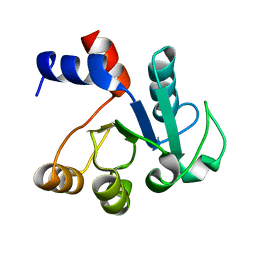 | | The structure of cell-free ID.343 from Thermus thermophilus | | Descriptor: | hypothetical protein TT1466 | | Authors: | Wada, T, Shirouzu, M, Park, S.-Y, Tame, J.R, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-05 | | Release date: | 2003-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a conserved CoA-binding protein synthesized by a cell-free system.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1J1V
 
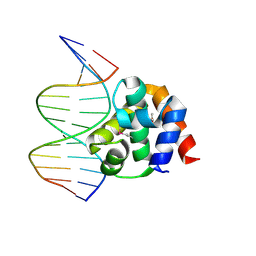 | | Crystal structure of DnaA domainIV complexed with DnaAbox DNA | | Descriptor: | 5'-D(*CP*CP*TP*GP*TP*GP*GP*AP*TP*AP*AP*CP*A)-3', 5'-D(*TP*GP*TP*TP*AP*TP*CP*CP*AP*CP*AP*GP*G)-3', Chromosomal replication initiator protein dnaA | | Authors: | Fujikawa, N, Kurumizaka, H, Nureki, O, Terada, T, Shirouzu, M, Katayama, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-18 | | Release date: | 2003-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of replication origin recognition by the DnaA protein
NUCLEIC ACIDS RES., 31, 2003
|
|
1WEH
 
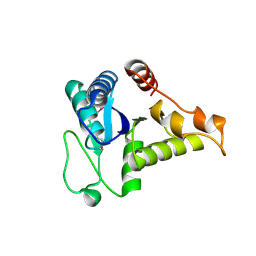 | | Crystal structure of the conserved hypothetical protein TT1887 from Thermus thermophilus HB8 | | Descriptor: | Conserved hypothetical protein TT1887 | | Authors: | Kukimoto-Niino, M, Murayama, K, Idaka, M, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of possible lysine decarboxylases from Thermus thermophilus HB8
PROTEIN SCI., 13, 2004
|
|
2YT8
 
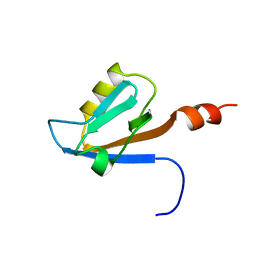 | | Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma) | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 3 | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Seiki, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma)
To be Published
|
|
1WEK
 
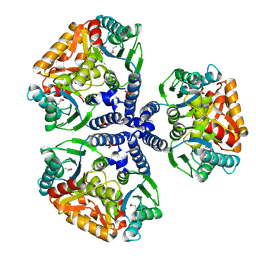 | | Crystal structure of the conserved hypothetical protein TT1465 from Thermus thermophilus HB8 | | Descriptor: | PHOSPHATE ION, hypothetical protein TT1465 | | Authors: | Kukimoto-Niino, M, Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of possible lysine decarboxylases from Thermus thermophilus HB8
Protein Sci., 13, 2004
|
|
2YT7
 
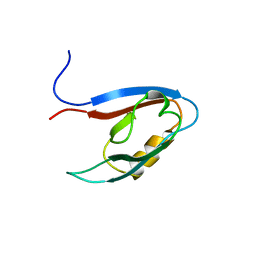 | | Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3 | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 3 | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Seiki, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3
To be Published
|
|
1WDV
 
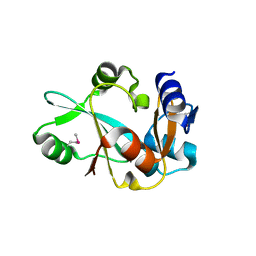 | | Crystal structure of hypothetical protein APE2540 | | Descriptor: | hypothetical protein APE2540 | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-18 | | Release date: | 2004-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a putative trans-editing enzyme for prolyl-tRNA synthetase from Aeropyrum pernix K1 at 1.7 A resolution.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2Z0T
 
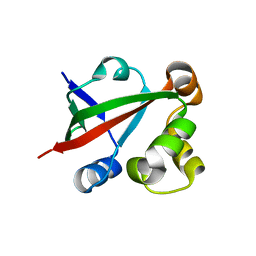 | | Crystal structure of hypothetical protein PH0355 | | Descriptor: | Putative uncharacterized protein PH0355 | | Authors: | Murayama, K, Takemoto, C, Kato-Murayama, M, Kawazoe, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hypothetical protein PH0355
To be Published
|
|
2ROK
 
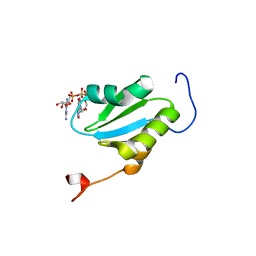 | | Solution structure of the cap-binding domain of PARN complexed with the cap analog | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, poly(A)-specific ribonuclease | | Authors: | Nagata, T, Suzuki, S, Endo, R, Shirouzu, M, Terada, T, Inoue, M, Kigawa, T, Guntert, P, Hayashizaki, Y, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-03-28 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The RRM domain of poly(A)-specific ribonuclease has a noncanonical binding site for mRNA cap analog recognition.
Nucleic Acids Res., 36, 2008
|
|
2ZDY
 
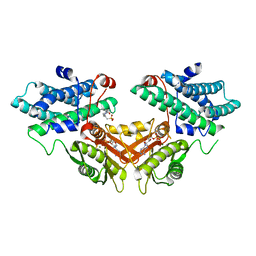 | | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kawamoto, M, Shiromizu, I, Kukimoto-Niino, M, Tokmakov, A, Terada, T, Shirouzu, M, Matsusue, T, Yokoyama, S. | | Deposit date: | 2007-12-01 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2RUG
 
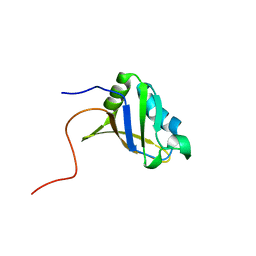 | | Refined solution structure of the first RNA recognition motif domain in CPEB3 | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 3 | | Authors: | Tsuda, K, Kuwasako, K, Nagata, T, Takahashi, M, Kigawa, T, Kobayashi, N, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2014-04-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel RNA recognition motif domain in the cytoplasmic polyadenylation element binding protein 3.
Proteins, 82, 2014
|
|
