1UGS
 
 | | Crystal structure of aY114T mutant of Co-type nitrile hydratase | | Descriptor: | COBALT (II) ION, Nitrile Hydratase alpha subunit, Nitrile Hydratase beta subunit | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Shoun, H, Wakagi, T. | | Deposit date: | 2003-06-17 | | Release date: | 2004-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding
Eur.J.Biochem., 271, 2004
|
|
5XB7
 
 | | GH42 alpha-L-arabinopyranosidase from Bifidobacterium animalis subsp. lactis Bl-04 | | Descriptor: | Beta-galactosidase, GLYCEROL, SULFATE ION | | Authors: | Viborg, A.H, Katayama, T, Arakawa, T, Abou Hachem, M, Lo Leggio, L, Kitaoka, M, Svensson, B, Fushinobu, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of alpha-l-arabinopyranosidases from human gut microbiome expands the diversity within glycoside hydrolase family 42.
J. Biol. Chem., 292, 2017
|
|
6K8Q
 
 | | Solution structure of the intermembrane space domain of the mitochondrial import protein Tim21 from S. cerevisiae | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM21 | | Authors: | Bala, S, Shinya, S, Srivastava, A, Shimada, A, Kobayashi, N, Kojima, C, Tama, F, Miyashita, O, Kohda, D. | | Deposit date: | 2019-06-13 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
5GKG
 
 | | Structure of EndoMS-dsDNA1'' complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*CP*AP*GP*GP*TP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*CP*GP*TP*GP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKJ
 
 | | Structure of EndoMS in apo form | | Descriptor: | Endonuclease EndoMS | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKF
 
 | | Structure of EndoMS-dsDNA1' complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*CP*AP*TP*GP*TP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*CP*TP*TP*GP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKE
 
 | | Structure of EndoMS-dsDNA1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*CP*AP*TP*GP*TP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*CP*GP*TP*GP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKH
 
 | | Structure of EndoMS-dsDNA2 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*CP*GP*GP*CP*AP*CP*TP*TP*GP*GP*CP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*GP*CP*CP*AP*GP*GP*TP*GP*CP*CP*GP*T)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKI
 
 | | Structure of EndoMS-dsDNA3 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*CP*CP*TP*AP*GP*GP*TP*CP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*GP*GP*GP*CP*CP*TP*AP*GP*GP*C)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
7BZL
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol | | Descriptor: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | Deposit date: | 2020-04-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
2D43
 
 | | Crystal structure of arabinofuranosidase complexed with arabinotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Miwa, Y, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2005-10-07 | | Release date: | 2006-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The family 42 carbohydrate-binding module of family 54 alpha-L-arabinofuranosidase specifically binds the arabinofuranose side chain of hemicellulose
Biochem.J., 399, 2006
|
|
6KPO
 
 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris D154N/E156Q mutant in complex with fucosyl-N-acetylglucosamine-Asn | | Descriptor: | ASPARAGINE, Chitinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
6KPN
 
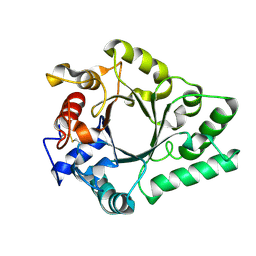 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris D154N/E156Q mutant in complex with fucosyl-N-acetylglucosamine | | Descriptor: | Chitinase, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
6KPM
 
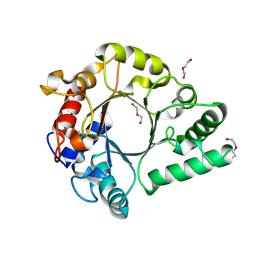 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in complex with L-fucose | | Descriptor: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
6KPL
 
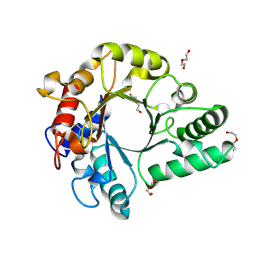 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in apo form | | Descriptor: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
4WH2
 
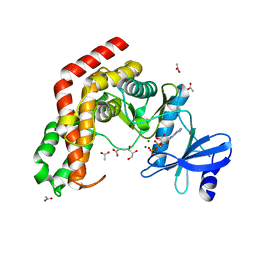 | | N-acetylhexosamine 1-kinase in complex with ADP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WH1
 
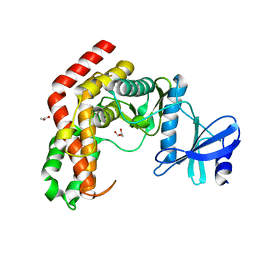 | | N-Acetylhexosamine 1-kinase (ligand free) | | Descriptor: | ACETIC ACID, GLYCEROL, N-acetylhexosamine 1-kinase | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WH3
 
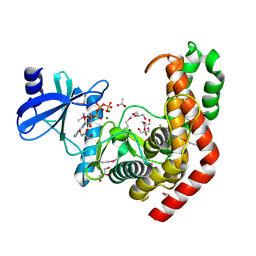 | | N-acetylhexosamine 1-kinase in complex with ATP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
1IZ4
 
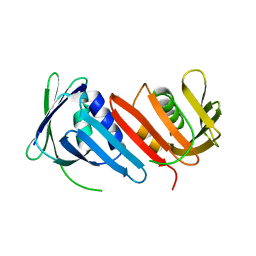 | |
6JI2
 
 | | Crystal structure of archaeal ribosomal protein aP1, aPelota, and GTP-bound aEF1A complex | | Descriptor: | Archaeal ribosomal stalk protein aP1, Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Maruyama, K, Imai, H, Kawamura, M, Ishino, S, Ishino, Y, Ito, K, Uchiumi, T. | | Deposit date: | 2019-02-20 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Switch of the interactions between the ribosomal stalk and EF1A in the GTP- and GDP-bound conformations.
Sci Rep, 9, 2019
|
|
1ISQ
 
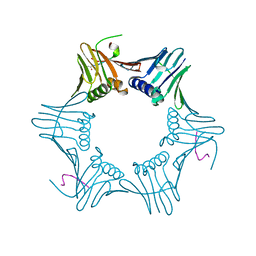 | | Pyrococcus furiosus PCNA complexed with RFCL PIP-box peptide | | Descriptor: | Proliferating Cell Nuclear Antigen, replication factor C large subunit | | Authors: | Matsumiya, S, Ishino, S, Ishino, Y, Morikawa, K. | | Deposit date: | 2001-12-19 | | Release date: | 2002-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Physical interaction between proliferating cell nuclear antigen and
replication factor C from Pyrococcus furiosus
Genes Cells, 7, 2002
|
|
1IZ5
 
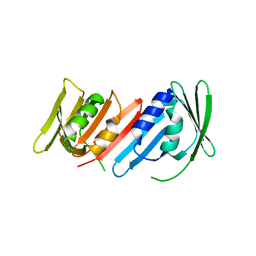 | | Pyrococcus furiosus PCNA mutant (Met73Leu, Asp143Ala, Asp147Ala): orthorhombic form | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Matsumiya, S, Ishino, S, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-09-23 | | Release date: | 2003-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Intermolecular ion pairs maintain the toroidal structure of Pyrococcus furiosus PCNA
PROTEIN SCI., 12, 2003
|
|
6KNC
 
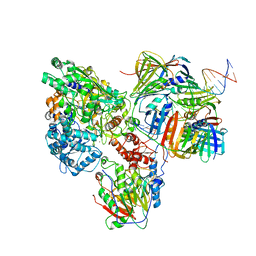 | | PolD-PCNA-DNA (form B) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
6KQT
 
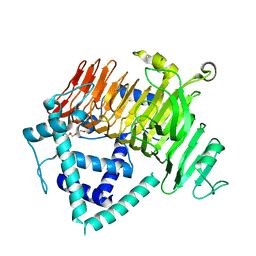 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein | | Descriptor: | SODIUM ION, TRIETHYLENE GLYCOL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
6KQS
 
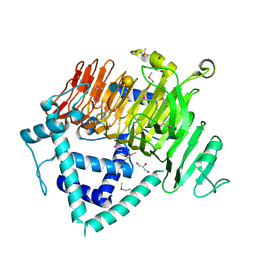 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative | | Descriptor: | GLYCEROL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, lacto-N-biosidase | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
