6KXE
 
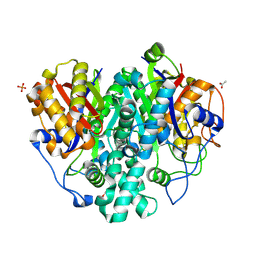 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Ketosynthase, ... | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
6KQS
 
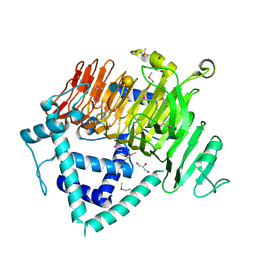 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative | | Descriptor: | GLYCEROL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, lacto-N-biosidase | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
3E5J
 
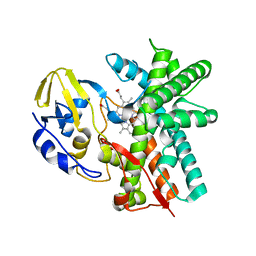 | | Crystal structure of CYP105P1 wild-type ligand-free form | | Descriptor: | Cytochrome P450 (Cytochrome P450 hydroxylase), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S, Ikeda, H, Wakagi, T, Shoun, H. | | Deposit date: | 2008-08-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state
J.Bacteriol., 191, 2009
|
|
3E5K
 
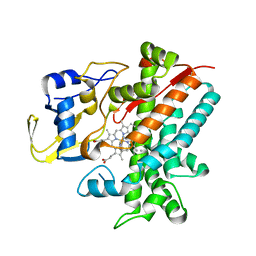 | | Crystal structure of CYP105P1 wild-type 4-phenylimidazole complex | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Cytochrome P450 (Cytochrome P450 hydroxylase), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S, Ikeda, H, Wakagi, T, Shoun, H. | | Deposit date: | 2008-08-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state
J.Bacteriol., 191, 2009
|
|
6KXF
 
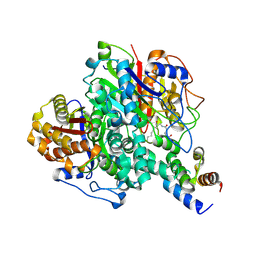 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | ACP, Ketosynthase, [(3~{R})-2,2-dimethyl-4-[[3-[2-[[(~{E})-oct-2-enoyl]amino]ethylamino]-3-oxidanylidene-propyl]amino]-3-oxidanyl-4-oxidanylidene-butyl] dihydrogen phosphate | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
6KXD
 
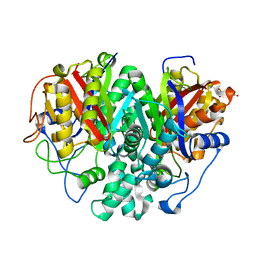 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Ketosynthase, ... | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
3E5L
 
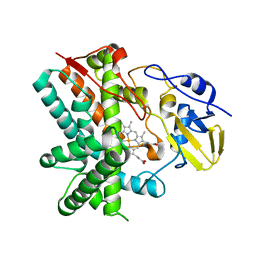 | | Crystal structure of CYP105P1 H72A mutant | | Descriptor: | Cytochrome P450 (Cytochrome P450 hydroxylase), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S, Ikeda, H, Wakagi, T, Shoun, H. | | Deposit date: | 2008-08-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state
J.Bacteriol., 191, 2009
|
|
6L69
 
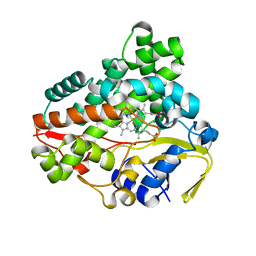 | | Crystal structure of CYP154C2 from Streptomyces avermitilis | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regio- and stereoselective hydroxylation of testosterone by a novel cytochrome P450 154C2 from Streptomyces avermitilis.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
5H40
 
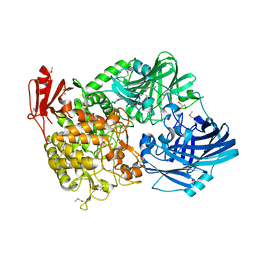 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose | | Descriptor: | CALCIUM ION, GLYCEROL, Uncharacterized protein, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
7V1W
 
 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium in complex with beta-D-arabinofuranose | | Descriptor: | CALCIUM ION, Difructose dianhydride I synthase/hydrolase (alphaFFase1), beta-D-arabinofuranose | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
7V1V
 
 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium, ligand-free form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, D(-)-TARTARIC ACID, ... | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
7V1X
 
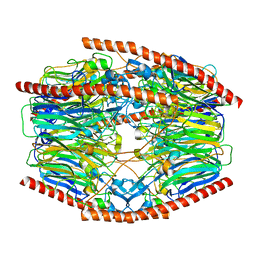 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium in complex with beta-D-fructofuranose | | Descriptor: | CALCIUM ION, Difructose dianhydride I synthase/hydrolase, beta-D-fructofuranose | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
8YK1
 
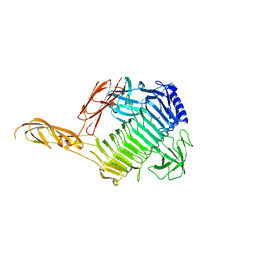 | | Blood group B alpha-1,3-galactosidase AgaBb from Bifidobacterium bifidum, construct 23-844 | | Descriptor: | Alpha-galactosidase, SODIUM ION | | Authors: | Kashima, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2024-03-04 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of Bifidobacterium bifidum Glycoside Hydrolase Family 110 alpha-Galactosidase Specific for Blood Group B Antigen.
J Appl Glycosci (1999), 71, 2024
|
|
6J9S
 
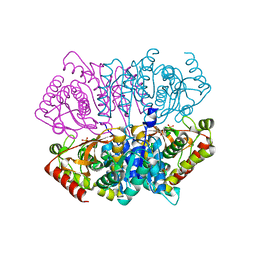 | | Penta mutant of Lactobacillus casei lactate dehydrogenase | | Descriptor: | GLYCEROL, L-lactate dehydrogenase, SULFATE ION | | Authors: | Arai, K, Miyanaga, A, Uchikoba, H, Fushinobu, S, Taguchi, H. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of penta mutant of L-lactate dehydrogenase from Lactobacillus casei
To Be Published
|
|
6J9T
 
 | | Complex structure of Lactobacillus casei lactate dehydrogenase with fructose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, L-lactate dehydrogenase, SULFATE ION | | Authors: | Arai, K, Miyanaga, A, Uchikoba, H, Fushinobu, S, Taguchi, H. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of penta mutant of L-lactate dehydrogenase from Lactobacillus casei
To Be Published
|
|
6J9U
 
 | | Complex structure of Lactobacillus casei lactate dehydrogenase penta mutant with pyruvate | | Descriptor: | L-lactate dehydrogenase, PYRUVIC ACID, SULFATE ION | | Authors: | Arai, K, Miyanaga, A, Uchikoba, H, Fushinobu, S, Taguchi, H. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of penta mutant of L-lactate dehydrogenase from Lactobacillus casei
To Be Published
|
|
3EQO
 
 | | Crystal structure of beta-1,3-glucanase from Phanerochaete chrysosporium (Lam55A) gluconolactone complex | | Descriptor: | D-glucono-1,5-lactone, Glucan 1,3-beta-glucosidase, ZINC ION, ... | | Authors: | Ishida, T, Fushinobu, S, Kawai, R, Kitaoka, M, Igarashi, K, Samejima, M. | | Deposit date: | 2008-10-01 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of glycoside hydrolase family 55 beta -1,3-glucanase from the basidiomycete Phanerochaete chrysosporium
J.Biol.Chem., 284, 2009
|
|
2MLO
 
 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a Membrane bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
8YK3
 
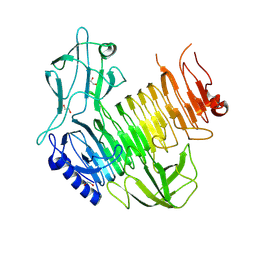 | | Blood group B alpha-1,3-galactosidase AgaBb from Bifidobacterium bifidum, construct T7-tag_24-673 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-galactosidase, ... | | Authors: | Kashima, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2024-03-04 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of Bifidobacterium bifidum Glycoside Hydrolase Family 110 alpha-Galactosidase Specific for Blood Group B Antigen.
J Appl Glycosci (1999), 71, 2024
|
|
8WU4
 
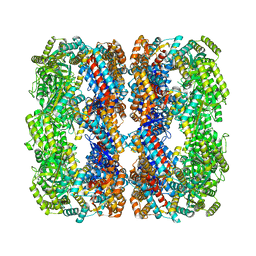 | | Cryo-EM structure of native H. thermoluteolus TH-1 GroEL | | Descriptor: | Chaperonin GroEL | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
5H42
 
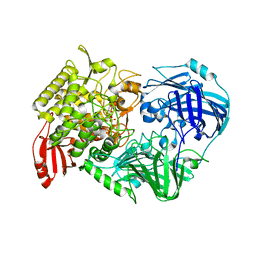 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with alpha-d-glucose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, Uncharacterized protein, alpha-D-glucopyranose | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
7WDT
 
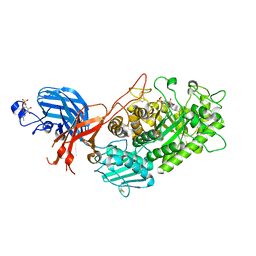 | | 6-sulfo-beta-D-N-acetylglucosaminidase from Bifidobacterium bifidum in complex with GlcNAc-6S | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, ... | | Authors: | Yamada, C, Kashima, T, Fushinobu, S, Katoh, T, Katayama, T. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem.
Nat.Chem.Biol., 19, 2023
|
|
7WDU
 
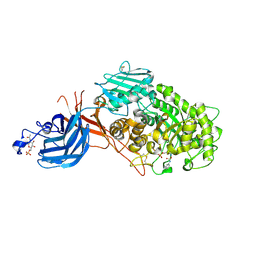 | | 6-sulfo-beta-D-N-acetylglucosaminidase from Bifidobacterium bifidum in complex with PUGNAc-6S | | Descriptor: | Beta-N-acetylhexosaminidase, CALCIUM ION, [[(3R,4R,5S,6R)-3-acetamido-4,5-bis(oxidanyl)-6-(sulfooxymethyl)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Kashima, T, Yamada, C, Fushinobu, S, Katoh, T, Katayama, T. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem.
Nat.Chem.Biol., 19, 2023
|
|
2MLQ
 
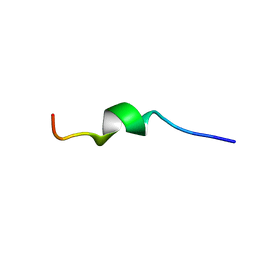 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a frount bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
1WD4
 
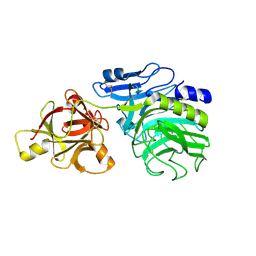 | | Crystal structure of arabinofuranosidase complexed with arabinose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2004-05-11 | | Release date: | 2004-09-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of a family 54 alpha-L-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose
J.Biol.Chem., 279, 2004
|
|
