2HYB
 
 | | Crystal Structure of Hexameric DsrEFH | | Descriptor: | DsrH, Intracellular sulfur oxidation protein dsrF, Putative sulfurtransferase dsrE | | Authors: | Shin, D.H, Connie, H, Schulte, A, Dahl, C, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-04 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hexameric DsrEFH
To be Published
|
|
2I15
 
 | |
2HY5
 
 | | Crystal structure of DsrEFH | | Descriptor: | DsrH, Intracellular sulfur oxidation protein dsrF, Putative sulfurtransferase dsrE | | Authors: | Shin, D.H, Schulte, A, Dahl, C, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of DsrEFH
To be Published
|
|
2I1O
 
 | | Crystal Structure of a Nicotinate Phosphoribosyltransferase from Thermoplasma acidophilum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Shin, D.H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-14 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a zinc ion bound Nicotinate Phosphoribosyltransferase from Thermoplasma acidophilum
To be Published
|
|
1T70
 
 | | Crystal structure of a novel phosphatase from Deinococcus radiodurans | | Descriptor: | Phosphatase | | Authors: | Shin, D.H, Wang, W, Kim, R, Yokota, H, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and enzymatic characterization of DR1281: A calcineurin-like phosphoesterase from Deinococcus radiodurans.
Proteins, 70, 2008
|
|
1NF2
 
 | |
1RQ0
 
 | | Crystal structure of peptide releasing factor 1 | | Descriptor: | Peptide chain release factor 1 | | Authors: | Shin, D.H, Brandsen, J, Jancarik, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-12-03 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome.
J.Mol.Biol., 341, 2004
|
|
1S12
 
 | | Crystal structure of TM1457 | | Descriptor: | ACETATE ION, hypothetical protein TM1457 | | Authors: | Shin, D.H, Lou, Y, Jancarik, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-05 | | Release date: | 2004-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of TM1457 from Thermotoga maritima.
J.Struct.Biol., 152, 2005
|
|
1T6X
 
 | | Crystal structure of ADP bound TM379 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, riboflavin kinase/FMN adenylyltransferase | | Authors: | Shin, D.H, Wang, W, Kim, R, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of ADP bound FAD synthetase
To be Published
|
|
1T6Z
 
 | | Crystal structure of riboflavin bound TM379 | | Descriptor: | RIBOFLAVIN, riboflavin kinase/FMN adenylyltransferase | | Authors: | Shin, D.H, Wang, W, Kim, R, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ADP bound FAD synthetase
To be Published
|
|
1T6Y
 
 | | Crystal structure of ADP, AMP, and FMN bound TM379 | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Shin, D.H, Wang, W, Kim, R, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ADP bound FAD synthetase
To be Published
|
|
1T71
 
 | | Crystal structure of a novel phosphatase Mycoplasma pneumoniaefrom | | Descriptor: | FE (III) ION, phosphatase | | Authors: | Shin, D.H, Jancarik, J, Kim, R, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a novel phosphatase from Mycoplasma pneumoniae
To be Published
|
|
1U0L
 
 | | Crystal structure of YjeQ from Thermotoga maritima | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Probable GTPase engC, ZINC ION | | Authors: | Shin, D.H, Lou, Y, Jaru, J, Kim, R, Yokota, H, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-07-13 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of YjeQ from Thermotoga maritima contains a circularly permuted GTPase domain
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1L2F
 
 | | Crystal structure of NusA from Thermotoga maritima: a structure-based role of the N-terminal domain | | Descriptor: | N utilization substance protein A | | Authors: | Shin, D.H, Nguyen, H.H, Jancarik, J, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-02-20 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of NusA from Thermotoga maritima and functional implication of the N-terminal domain.
Biochemistry, 42, 2003
|
|
1LFP
 
 | | Crystal Structure of a Conserved Hypothetical Protein Aq1575 from Aquifex Aeolicus | | Descriptor: | Hypothetical protein AQ_1575 | | Authors: | Shin, D.H, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-04-11 | | Release date: | 2002-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of conserved hypothetical protein Aq1575 from Aquifex aeolicus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2O5K
 
 | | Crystal Structure of GSK3beta in complex with a benzoimidazol inhibitor | | Descriptor: | 2-(2,4-DICHLORO-PHENYL)-7-HYDROXY-1H-BENZOIMIDAZOLE-4-CARBOXYLIC ACID [2-(4-METHANESULFONYLAMINO-PHENYL)-ETHYL]-AMIDE, Glycogen synthase kinase-3 beta | | Authors: | Shin, D, Lee, S.C, Heo, Y.S, Cho, Y.S, Kim, Y.E, Hyun, Y.L, Cho, J.M, Lee, Y.S, Ro, S. | | Deposit date: | 2006-12-06 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 7-hydroxy-1H-benzoimidazole derivatives as novel inhibitors of glycogen synthase kinase-3beta
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4V46
 
 | | Crystal structure of the BAFF-BAFF-R complex | | Descriptor: | MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 13B, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Kim, H.M, Yu, K.S, Lee, M.E, Shin, D.R, Kim, Y.S, Paik, S.G, Yoo, O.J, Lee, H, Lee, J.-O. | | Deposit date: | 2003-03-23 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation
NAT.STRUCT.BIOL., 10, 2003
|
|
5JXD
 
 | | Crystal structure of murine Tnfaip8 C165S mutant | | Descriptor: | Tumor necrosis factor alpha-induced protein 8, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Park, J, Kim, M.S, Lee, D, Shin, D.H. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | The Tnfaip8-PE complex is a novel upstream effector in the anti-autophagic action of insulin
Sci Rep, 7, 2017
|
|
4Q9D
 
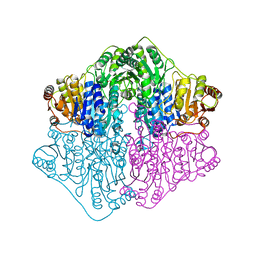 | | X-ray structure of a putative thiamin diphosphate-dependent enzyme isolated from Mycobacterium smegmatis | | Descriptor: | Benzoylformate decarboxylase, FORMIC ACID, MAGNESIUM ION | | Authors: | Andrews, F.H, Horton, J.D, Yoon, H.J, Malik, A.M.K, Logsdon, M.G, Shin, D.H, Kneen, M.M, Suh, S.W, McLeish, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The kinetic characterization and X-ray structure of a putative benzoylformate decarboxylase from M. smegmatis highlights the difficulties in the functional annotation of ThDP-dependent enzymes.
Biochim.Biophys.Acta, 1854, 2015
|
|
3JRW
 
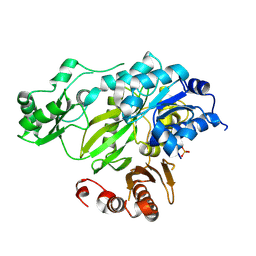 | | Phosphorylated BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3PJF
 
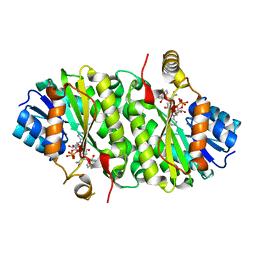 | |
3JRX
 
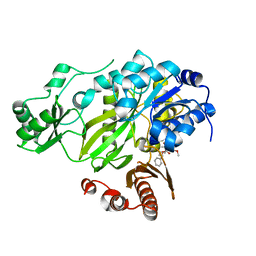 | | Crystal structure of the BC domain of ACC2 in complex with soraphen A | | Descriptor: | Acetyl-CoA carboxylase 2, SORAPHEN A | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
4PNH
 
 | |
3DSC
 
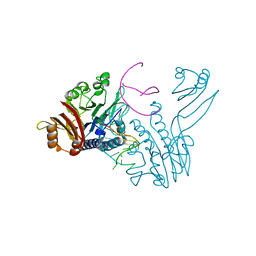 | | Crystal structure of P. furiosus Mre11 DNA synaptic complex | | Descriptor: | DNA (5'-D(P*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DAP*DC)-3'), DNA double-strand break repair protein mre11 | | Authors: | Williams, R.S, Moncalian, G, Shin, D.S, Tainer, J.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
3PJE
 
 | |
