2ZLY
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
2ZM9
 
 | | Structure of 6-Aminohexanoate-dimer Hydrolase, A61V/S112A/A124V/R187S/F264C/G291R/G338A/D370Y mutant (Hyb-S4M94) with Substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
1KWH
 
 | | Structure Analysis AlgQ2, a Macromolecule(alginate)-Binding Periplasmic Protein of Sphingomonas sp. A1. | | Descriptor: | CALCIUM ION, Macromolecule-Binding Periplasmic Protein | | Authors: | Momma, K, Mikami, B, Mishima, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2002-01-29 | | Release date: | 2002-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of AlgQ2, a macromolecule (alginate)-binding protein of Sphingomonas sp. A1 at 2.0A resolution.
J.Mol.Biol., 316, 2002
|
|
2ZD0
 
 | | Crystal structures and thermostability of mutant TRAP3 A5 (ENGINEERED TRAP) | | Descriptor: | TRYPTOPHAN, Transcription attenuation protein mtrB | | Authors: | Watanabe, M, Mishima, Y, Yamashita, I, Park, S.Y, Tame, J.R.H, Heddle, J.G. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intersubunit linker length as a modifier of protein stability: crystal structures and thermostability of mutant TRAP.
Protein Sci., 17, 2008
|
|
2ZM2
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, A61V/A124V/R187S/F264C/G291R/G338A/D370Y mutant (Hyb-S4M94) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Nego, S. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
7F4Z
 
 | | X-ray crystal structure of Y149A mutated Hsp72-NBD in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Heat shock 70 kDa protein 1B, ... | | Authors: | Yokoyama, T, Fujii, S, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
7F50
 
 | | X-ray crystal structure of Y149A mutated Hsp72-NBD in complex with AMPPnP | | Descriptor: | CHLORIDE ION, Heat shock 70 kDa protein 1B, MAGNESIUM ION, ... | | Authors: | Yokoyama, T, Fujii, S, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
3WJ2
 
 | | Crystal structure of ESTFA (FE-lacking apo form) | | Descriptor: | Carboxylesterase | | Authors: | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | Deposit date: | 2013-10-03 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
1V29
 
 | | Crystal structure of Nitrile hydratase from a thermophile Bacillus smithii | | Descriptor: | COBALT (II) ION, nitrile hydratase a chain, nitrile hydratase b chain | | Authors: | Hourai, S, Miki, M, Takashima, Y, Mitsuda, S, Yanagi, K. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of nitrile hydratase from a thermophilic Bacillus smithii
Biochem.Biophys.Res.Commun., 312, 2003
|
|
4CDO
 
 | | Crystal structure of PQBP1 bound to spliceosomal U5-15kD | | Descriptor: | THIOREDOXIN-LIKE PROTEIN 4A, POLYGLUTAMINE-BINDING PROTEIN | | Authors: | Mizuguchi, M, Obita, T, Serita, T, Kojima, R, Nabeshima, Y, Okazawa, H. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutations in the Pqbp1 Gene Prevent its Interaction with the Spliceosomal Protein U5-15Kd.
Nat.Commun., 5, 2014
|
|
4BWS
 
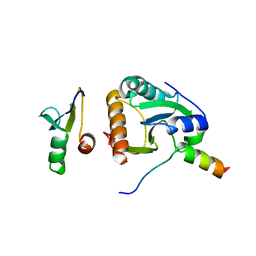 | | Crystal structure of the heterotrimer of PQBP1, U5-15kD and U5-52kD. | | Descriptor: | CD2 ANTIGEN CYTOPLASMIC TAIL-BINDING PROTEIN 2, POLYGLUTAMINE-BINDING PROTEIN 1, THIOREDOXIN-LIKE PROTEIN 4A | | Authors: | Mizuguchi, M, Obita, T, Serita, T, Kojima, R, Morimoto, T, Nabeshima, Y, Okazawa, H. | | Deposit date: | 2013-07-04 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutations in the Pqbp1 Gene Prevent its Interaction with the Spliceosomal Protein U5-15Kd.
Nat.Commun., 5, 2014
|
|
3A5P
 
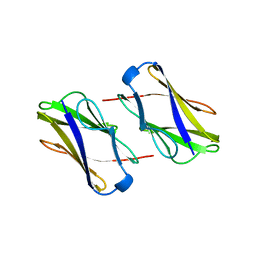 | | Crystal structure of hemagglutinin | | Descriptor: | Haemagglutinin I | | Authors: | Watanabe, N, Sakai, N, Nakamura, T, Nabeshima, Y, Kouno, T, Mizuguchi, M, Kawano, K. | | Deposit date: | 2009-08-10 | | Release date: | 2010-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Structure of Physarum polycephalum hemagglutinin I suggests a minimal carbohydrate recognition domain of legume lectin fold
J.Mol.Biol., 405, 2011
|
|
2E8J
 
 | |
