2RVB
 
 | |
2RNF
 
 | | X-RAY CRYSTAL STRUCTURE OF HUMAN RIBONUCLEASE 4 IN COMPLEX WITH D(UP) | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, RIBONUCLEASE 4 | | Authors: | Terzyan, S.S, Peracaula, R, De Llorens, R, Tsushima, Y, Yamada, H, Seno, M, Gomis-Ruth, F.X, Coll, M. | | Deposit date: | 1998-11-03 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of human RNase 4, unliganded and complexed with d(Up), reveals the basis for its uridine selectivity.
J.Mol.Biol., 285, 1999
|
|
2RUK
 
 | |
2RVN
 
 | |
2RVL
 
 | |
6K5W
 
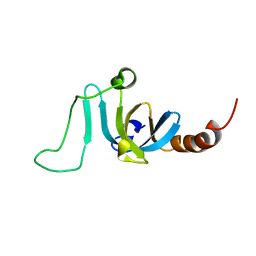 | | Solution structure of the chromodomain of yeast Eaf3 | | Descriptor: | Chromatin modification-related protein EAF3 | | Authors: | Okuda, M, Nishimura, Y. | | Deposit date: | 2019-05-31 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Eaf3 chromodomain acts as a pH sensor for gene expression by altering its binding affinity for histone methylated-lysine residues.
Biosci.Rep., 40, 2020
|
|
1GEB
 
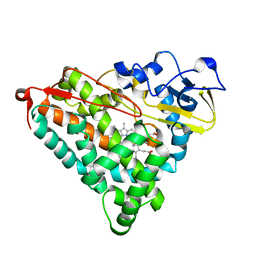 | | X-RAY CRYSTAL STRUCTURE AND CATALYTIC PROPERTIES OF THR252ILE MUTANT OF CYTOCHROME P450CAM | | Descriptor: | CAMPHOR, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hishiki, T, Shimada, H, Nagano, S, Park, S.-Y, Ishimura, Y. | | Deposit date: | 2000-11-01 | | Release date: | 2000-11-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | X-ray crystal structure and catalytic properties of Thr252Ile mutant of cytochrome P450cam: roles of Thr252 and water in the active center.
J.Biochem., 128, 2000
|
|
3WJ1
 
 | | Crystal structure of SSHESTI | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | Deposit date: | 2013-10-03 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
1MBH
 
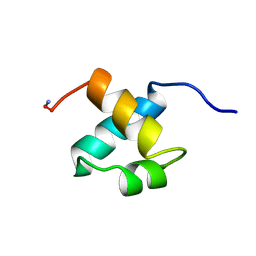 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | C-MYB | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
5YK9
 
 | |
2LX3
 
 | | 1H,13C,15N assignments for an isoform of the type III antifreeze protein from notched-fin eelpout | | Descriptor: | Type III antifreeze protein nfeAFP11 | | Authors: | Kumeta, H, Ogura, K, Nishimiya, Y, Miura, A, Inagaki, F, Tsuda, S. | | Deposit date: | 2012-08-12 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure note: a defective isoform and its activity-improved variant of a type III antifreeze protein from Zoarces elongates Kner
J.Biomol.Nmr, 55, 2013
|
|
2LX2
 
 | | 1H,13C,15N assignments for an isoform of the type III antifreeze protein from notched-fin eelpout | | Descriptor: | Type III antifreeze protein nfeAFP11 | | Authors: | Kumeta, H, Ogura, K, Nishimiya, Y, Miura, A, Inagaki, F, Tsuda, S. | | Deposit date: | 2012-08-12 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure note: a defective isoform and its activity-improved variant of a type III antifreeze protein from Zoarces elongates Kner
J.Biomol.Nmr, 55, 2013
|
|
3V10
 
 | | Crystal structure of the collagen binding domain of Erysipelothrix rhusiopathiae surface protein RspB | | Descriptor: | Rhusiopathiae surface protein B | | Authors: | Ponnuraj, K, Swarmistha devi, A, Ogawa, Y, Shimoji, Y, Subramainan, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-10-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Collagen adhesin-nanoparticle interaction impairs adhesin's ligand binding mechanism
Biochim.Biophys.Acta, 1820, 2012
|
|
3WP9
 
 | | Crystal structure of antifreeze protein from an Antarctic sea ice bacterium Colwellia sp. | | Descriptor: | Ice-binding protein | | Authors: | Hanada, Y, Nishimiya, Y, Miura, A, Tsuda, S, Kondo, H. | | Deposit date: | 2014-01-10 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hyperactive antifreeze protein from an Antarctic sea ice bacterium Colwellia sp. has a compound ice-binding site without repetitive sequences.
Febs J., 281, 2014
|
|
5XV8
 
 | |
2ZM8
 
 | | Structure of 6-Aminohexanoate-dimer Hydrolase, S112A/D370Y Mutant Complexed with 6-Aminohexanoate-dimer | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
1IWK
 
 | | Putidaredoxin-Binding Stablilizes an Active Conformer of Cytochrome P450cam in its Reduced State; Crystal Structure of Mutant(112K) Cytochrome P450cam | | Descriptor: | CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Shimada, H, Tarumi, A, Hishiki, T, Kimata-Ariga, Y, Egawa, T, Park, S.-Y, Adachi, S, Shiro, Y, Ishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-05-15 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infrared spectroscopic and mutational studies on putidaredoxin-induced conformational changes in ferrous CO-P450cam
Biochemistry, 42, 2003
|
|
2ZM2
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, A61V/A124V/R187S/F264C/G291R/G338A/D370Y mutant (Hyb-S4M94) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Nego, S. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
2ZLY
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
2ZM9
 
 | | Structure of 6-Aminohexanoate-dimer Hydrolase, A61V/S112A/A124V/R187S/F264C/G291R/G338A/D370Y mutant (Hyb-S4M94) with Substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
1IWI
 
 | | Putidaredoxin-Binding Stablilizes an Active Conformer of Cytochrome P450cam in its Reduced State; Crystal Structure of Cytochrome P450cam | | Descriptor: | CAMPHOR, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Shimada, H, Tarumi, A, Hishiki, T, Kimata-Ariga, Y, Egawa, T, Park, S.-Y, Adachi, S, Shiro, Y, Ishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-05-15 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infrared spectroscopic and mutational studies on putidaredoxin-induced conformational changes in ferrous CO-P450cam
Biochemistry, 42, 2003
|
|
1IWJ
 
 | | Putidaredoxin-Binding Stablilizes an Active Conformer of Cytochrome P450cam in its Reduced State; Crystal Structure of Mutant(109K) Cytochrome P450cam | | Descriptor: | CAMPHOR, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Shimada, H, Tarumi, A, Hishiki, T, Kimata-Ariga, Y, Egawa, T, Park, S.-Y, Adachi, S, Shiro, Y, Ishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-05-15 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infrared spectroscopic and mutational studies on putidaredoxin-induced conformational changes in ferrous CO-P450cam
Biochemistry, 42, 2003
|
|
2ZD0
 
 | | Crystal structures and thermostability of mutant TRAP3 A5 (ENGINEERED TRAP) | | Descriptor: | TRYPTOPHAN, Transcription attenuation protein mtrB | | Authors: | Watanabe, M, Mishima, Y, Yamashita, I, Park, S.Y, Tame, J.R.H, Heddle, J.G. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intersubunit linker length as a modifier of protein stability: crystal structures and thermostability of mutant TRAP.
Protein Sci., 17, 2008
|
|
8H8Q
 
 | | Fab-amyloid beta fragment complex at neutral pH | | Descriptor: | CHLORIDE ION, Fab, GLN-LYS-CYS-VAL-PHE-PHE-ALA-GLU-ASP-VAL-GLY-SER-ASN-CYS-GLY, ... | | Authors: | Kita, A, Irie, K, Irie, Y, Matsushima, Y, Miki, K. | | Deposit date: | 2022-10-24 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fab-amyloid beta fragment complex at neutral pH
To Be Published
|
|
1MSF
 
 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | Descriptor: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-01-24 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
