6AJW
 
 | | Crystal structure of BRD4 in complex with DMSO (Cocktail No. 4) | | Descriptor: | Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, SODIUM ION | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
5C5N
 
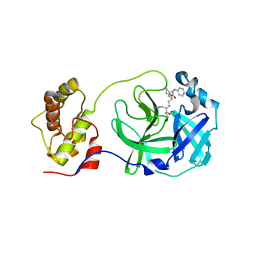 | | Structure of SARS-3CL protease complex with a phenyl-beta-alanyl (R,S)-N-decalin type inhibitor | | Descriptor: | (2S)-3-(1H-imidazol-5-yl)-2-({[(3R,4aS,8aR)-2-(N-phenyl-beta-alanyl)decahydroisoquinolin-3-yl]methyl}amino)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2015-06-21 | | Release date: | 2016-06-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fused-ring structure of N-decalin as a novel scaffold for SARS 3CL protease inhibitors
to be published
|
|
1JF6
 
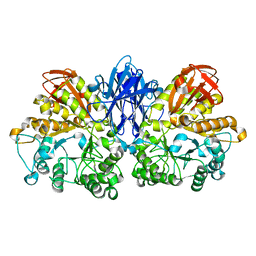 | | Crystal structure of thermoactinomyces vulgaris r-47 alpha-amylase mutant F286Y | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
1JF5
 
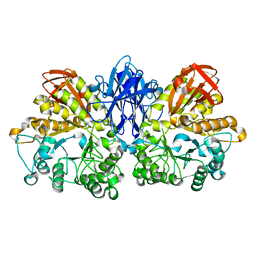 | | CRYSTAL STRUCTURE OF THERMOACTINOMYCES VULGARIS R-47 ALPHA-AMYLASE 2 MUTANT F286A | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
1JIB
 
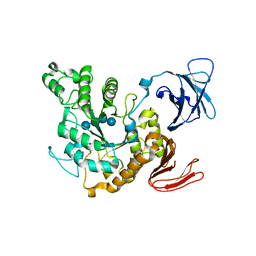 | | Complex of Alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with Maltotetraose Based on a Crystal Soaked with Maltohexaose. | | Descriptor: | NEOPULLULANASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2001-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
1QQI
 
 | | SOLUTION STRUCTURE OF THE DNA-BINDING AND TRANSACTIVATION DOMAIN OF PHOB FROM ESCHERICHIA COLI | | Descriptor: | PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Okamura, H, Hanaoka, S, Nagadoi, A, Makino, K, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-06-07 | | Release date: | 2000-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box.
J.Mol.Biol., 295, 2000
|
|
1VD4
 
 | | Solution structure of the zinc finger domain of TFIIE alpha | | Descriptor: | Transcription initiation factor IIE, alpha subunit, ZINC ION | | Authors: | Okuda, M, Tanaka, A, Arai, Y, Satoh, M, Okamura, H, Nagadoi, A, Hanaoka, F, Ohkuma, Y, Nishimura, Y. | | Deposit date: | 2004-03-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc finger structure in the large subunit of human general transcription factor TFIIE.
J.Biol.Chem., 279, 2004
|
|
1WZK
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt D465N | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1WZL
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt R469L | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
2RVM
 
 | |
1WZM
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt R469K | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
2RVB
 
 | |
8I53
 
 | |
2RNF
 
 | | X-RAY CRYSTAL STRUCTURE OF HUMAN RIBONUCLEASE 4 IN COMPLEX WITH D(UP) | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, RIBONUCLEASE 4 | | Authors: | Terzyan, S.S, Peracaula, R, De Llorens, R, Tsushima, Y, Yamada, H, Seno, M, Gomis-Ruth, F.X, Coll, M. | | Deposit date: | 1998-11-03 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of human RNase 4, unliganded and complexed with d(Up), reveals the basis for its uridine selectivity.
J.Mol.Biol., 285, 1999
|
|
2RUK
 
 | |
2RVN
 
 | |
7F4Z
 
 | | X-ray crystal structure of Y149A mutated Hsp72-NBD in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Heat shock 70 kDa protein 1B, ... | | Authors: | Yokoyama, T, Fujii, S, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
7F50
 
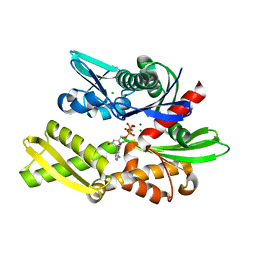 | | X-ray crystal structure of Y149A mutated Hsp72-NBD in complex with AMPPnP | | Descriptor: | CHLORIDE ION, Heat shock 70 kDa protein 1B, MAGNESIUM ION, ... | | Authors: | Yokoyama, T, Fujii, S, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
2RVL
 
 | |
1JL8
 
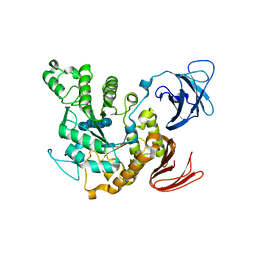 | | Complex of alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with beta-cyclodextrin based on a co-crystallization with methyl beta-cyclodextrin | | Descriptor: | ALPHA-AMYLASE II, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
5YK9
 
 | |
5C5O
 
 | | Structure of SARS-3CL protease complex with a phenyl-beta-alanyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-3-(1H-imidazol-5-yl)-2-({[(3S,4aR,8aS)-2-(N-phenyl-beta-alanyl)decahydroisoquinolin-3-yl]methyl}amino)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2015-06-21 | | Release date: | 2016-06-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fused-ring structure of N-decalin as a novel scaffold for SARS 3CL protease inhibitors
to be published
|
|
1UBB
 
 | | Crystal structure of rat HO-1 in complex with ferrous heme | | Descriptor: | Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Higashimoto, Y, Noguchi, M, Fukuyama, K. | | Deposit date: | 2003-04-03 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
5Y4G
 
 | | Apo Structure of AmbP3 | | Descriptor: | AmbP3 | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-03 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Y72
 
 | | DMSPP Bound AmbP3 | | Descriptor: | AmbP3, DIMETHYLALLYL S-THIOLODIPHOSPHATE | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
