3A0W
 
 | | Catalytic domain of histidine kinase ThkA (TM1359) for MAD phasing (nucleotide free form 2, orthorombic) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(ETHYLMERCURI-THIO)-BENZOIC ACID, ETHYL MERCURY ION, ... | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3A17
 
 | | Crystal Structure of Aldoxime Dehydratase (OxdRE) in Complex with Butyraldoxime (Co-crystal) | | Descriptor: | (1Z)-butanal oxime, Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
3A0X
 
 | | Catalytic domain of histidine kinase ThkA (TM1359) (nucleotide free form 1: ammomium phosphate, monoclinic) | | Descriptor: | Sensor protein | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-25 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3A15
 
 | | Crystal Structure of Substrate-Free Form of Aldoxime Dehydratase (OxdRE) | | Descriptor: | Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
3A0T
 
 | | Catalytic domain of histidine kinase ThkA (TM1359) in complex with ADP and Mg ion (trigonal) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Sensor protein | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3A0Z
 
 | | Catalytic domain of histidine kinase ThkA (TM1359) (nucleotide free form 4: isopropanol, orthorombic) | | Descriptor: | Sensor protein | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-25 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3AGF
 
 | |
3ANW
 
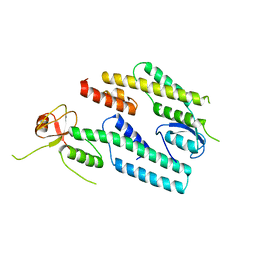 | | A protein complex essential initiation of DNA replication | | Descriptor: | Putative uncharacterized protein | | Authors: | Oyama, T, Shirai, T, Nagasawa, N, Ishino, Y, Morikawa, K. | | Deposit date: | 2010-09-10 | | Release date: | 2011-07-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Architectures of archaeal GINS complexes, essential DNA replication initiation factors
Bmc Biol., 9, 2011
|
|
3AJ3
 
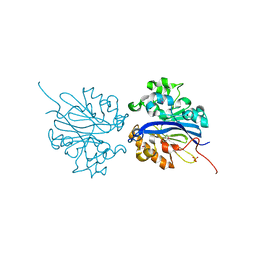 | | Crystal structure of selenomethionine substituted 4-pyridoxolactonase from Mesorhizobium loti | | Descriptor: | 4-pyridoxolactonase, PHOSPHATE ION, ZINC ION | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mikami, B, Yagi, T. | | Deposit date: | 2010-05-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.577 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3WD8
 
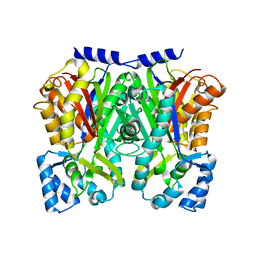 | | TypeIII polyketide synthases | | Descriptor: | GLYCEROL, Type III polyketide synthase quinolone synthase | | Authors: | Mori, T, Shimokawa, Y, Matsui, T, Morita, H, Abe, I. | | Deposit date: | 2013-06-10 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | Cloning, characterization, and crystal structure analysis of novel type III polyketide synthases from Citrus microcarpa
To be Published
|
|
3WI3
 
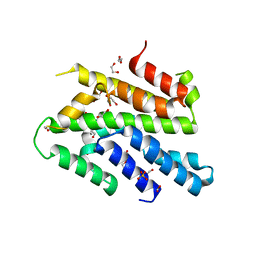 | | Crystal Structure of the Sld3/Treslin domain from yeast Sld3 | | Descriptor: | 1,2-ETHANEDIOL, DNA replication regulator SLD3, SULFATE ION | | Authors: | Itou, H, Araki, H, Shirakihara, Y. | | Deposit date: | 2013-09-05 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the homology domain of the eukaryotic DNA replication proteins sld3/treslin.
Structure, 22, 2014
|
|
3WN6
 
 | | Crystal structure of alpha-amylase AmyI-1 from Oryza sativa | | Descriptor: | Alpha-amylase, CALCIUM ION, D(-)-TARTARIC ACID, ... | | Authors: | Ochiai, A, Sugai, H, Harada, K, Tanaka, S, Ishiyama, Y, Ito, K, Tanaka, T, Uchiumi, T, Taniguchi, M, Mitsui, T. | | Deposit date: | 2013-12-05 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of alpha-amylase from Oryza sativa: molecular insights into enzyme activity and thermostability
Biosci.Biotechnol.Biochem., 78, 2014
|
|
3B1E
 
 | | Crystal structure of betaC-S lyase from Streptococcus anginosus in complex with L-serine: alpha-Aminoacrylate form | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Kezuka, Y, Yoshida, Y, Nonaka, T. | | Deposit date: | 2011-06-29 | | Release date: | 2012-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural insights into catalysis by beta C-S lyase from Streptococcus anginosus
Proteins, 80, 2012
|
|
3AQB
 
 | | M. luteus B-P 26 heterodimeric hexaprenyl diphosphate synthase in complex with magnesium | | Descriptor: | CHLORIDE ION, Component A of hexaprenyl diphosphate synthase, Component B of hexaprenyl diphosphate synthase, ... | | Authors: | Sasaki, D, Fujihashi, M, Okuyama, N, Kobayashi, Y, Noike, M, Koyama, T, Miki, K. | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of heterodimeric hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26 reveals that the small subunit is directly involved in the product chain length regulation.
J.Biol.Chem., 286, 2011
|
|
3B1C
 
 | | Crystal structure of betaC-S lyase from Streptococcus anginosus: Internal aldimine form | | Descriptor: | BetaC-S lyase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Kezuka, Y, Yoshida, Y, Nonaka, T. | | Deposit date: | 2011-06-29 | | Release date: | 2012-06-27 | | Last modified: | 2014-01-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural insights into catalysis by beta C-S lyase from Streptococcus anginosus
Proteins, 80, 2012
|
|
3B1D
 
 | | Crystal structure of betaC-S lyase from Streptococcus anginosus in complex with L-serine: External aldimine form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BetaC-S lyase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Kezuka, Y, Yoshida, Y, Nonaka, T. | | Deposit date: | 2011-06-29 | | Release date: | 2012-06-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural insights into catalysis by beta C-S lyase from Streptococcus anginosus
Proteins, 80, 2012
|
|
4AQ5
 
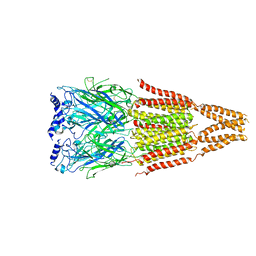 | |
4AQ9
 
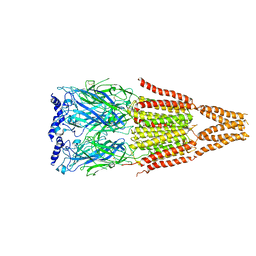 | |
3X37
 
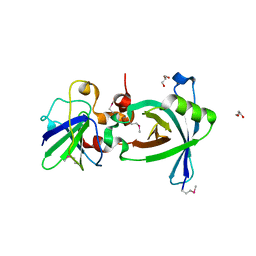 | | Crystal structure of the N-terminal domain of Sld7 in complex with Sld3 | | Descriptor: | GLYCEROL, Mitochondrial morphogenesis protein SLD7, ZYRO0C14696p | | Authors: | Itou, H, Araki, H, Shirakihara, Y. | | Deposit date: | 2015-01-16 | | Release date: | 2015-08-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The quaternary structure of the eukaryotic DNA replication proteins Sld7 and Sld3.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2ZOX
 
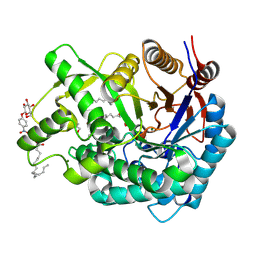 | | Crystal Structure of the Covalent Intermediate of Human Cytosolic beta-Glucosidase | | Descriptor: | 4-nitrophenyl alpha-D-glucopyranoside, Cytosolic beta-glucosidase, GLYCEROL, ... | | Authors: | Noguchi, J, Hayashi, Y, Baba, Y, Okino, N, Kimura, M, Ito, M, Kakuta, Y. | | Deposit date: | 2008-06-17 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the covalent intermediate of human cytosolic beta-glucosidase
Biochem.Biophys.Res.Commun., 374, 2008
|
|
3AQC
 
 | | M. luteus B-P 26 heterodimeric hexaprenyl diphosphate synthase in complex with magnesium and FPP analogue | | Descriptor: | (2E,6E)-7,11-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, CHLORIDE ION, Component A of hexaprenyl diphosphate synthase, ... | | Authors: | Sasaki, D, Fujihashi, M, Okuyama, N, Kobayashi, Y, Noike, M, Koyama, T, Miki, K. | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of heterodimeric hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26 reveals that the small subunit is directly involved in the product chain length regulation.
J.Biol.Chem., 286, 2011
|
|
3AYF
 
 | | Crystal structure of nitric oxide reductase | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Matsumoto, Y, Tosha, T, Pisliakov, A.V, Hino, T, Sugimoti, H, Nagano, S, Sugita, Y, Shiro, Y. | | Deposit date: | 2011-05-06 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of quinol-dependent nitric oxide reductase from Geobacillus stearothermophilus
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AYG
 
 | | Crystal structure of nitric oxide reductase complex with HQNO | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, CALCIUM ION, ... | | Authors: | Matsumoto, Y, Tosha, T, Pisliakov, A.V, Hino, T, Sugimoti, H, Nagano, S, Sugita, Y, Shiro, Y. | | Deposit date: | 2011-05-06 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of quinol-dependent nitric oxide reductase from Geobacillus stearothermophilus.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3WV5
 
 | | Complex structure of VinN with 3-methylaspartate | | Descriptor: | (2S,3S)-3-methyl-aspartic acid, Non-ribosomal peptide synthetase | | Authors: | Miyanaga, A, Cieslak, J, Shinohara, Y, Kudo, F, Eguchi, T. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the adenylation enzyme VinN reveals a unique beta-amino acid recognition mechanism
J.Biol.Chem., 289, 2014
|
|
2D4P
 
 | | Crystal structure of TTHA1254 (wild type) from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TTHA1254 | | Authors: | Mizohata, E, Uchikubo, T, Kinoshita, Y, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-21 | | Release date: | 2006-04-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of TTHA1254 (wild type) from Thermus thermophilus HB8
To be Published
|
|
