8JOG
 
 | | solution structure of Ras Binding Domein (RBD) in C-RAF with negative allosteric modulator. | | Descriptor: | RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Makino, Y, Matsumoto, S, Yoshikawa, Y, Kawamura, T, Kumasaka, T, Shima, F. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Small-molecule RAS/RAF-binding Inhibitors Allosterically Disrupt RAF Conformation and Exert Efficacy Against Broad-spectrum RAS-driven Cancers
To Be Published
|
|
6JLG
 
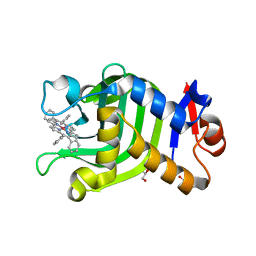 | | Crystal Structure of HasAp with Co-9,10,19,20-Tetraphenylporphycene | | Descriptor: | GLYCEROL, Heme acquisition protein HasA, PHOSPHATE ION, ... | | Authors: | Sakakibara, E, Shisaka, Y, Onoda, H, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-03-05 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly malleable haem-binding site of the haemoprotein HasA permits stable accommodation of bulky tetraphenylporphycenes.
Rsc Adv, 9, 2019
|
|
5AX6
 
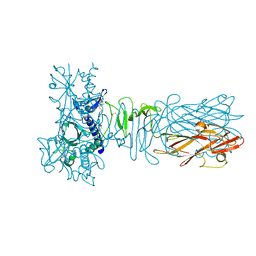 | | The crystal structure of CofB, the minor pilin subunit of CFA/III from human enterotoxigenic Escherichia coli. | | Descriptor: | ACETATE ION, CofB | | Authors: | Kawahara, K, Oki, K, Fukaksua, F, Maruno, T, Kobayashi, Y, Daisuke, M, Taniguchi, T, Honda, T, Iida, T, Nakamura, S, Ohkubo, T. | | Deposit date: | 2015-07-16 | | Release date: | 2016-03-09 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Homo-trimeric Structure of the Type IVb Minor Pilin CofB Suggests Mechanism of CFA/III Pilus Assembly in Human Enterotoxigenic Escherichia coli
J.Mol.Biol., 428, 2016
|
|
5AYJ
 
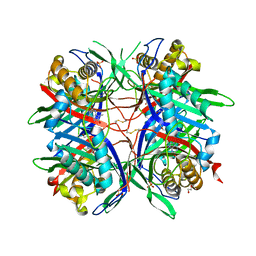 | | Hyperthermostable mutant of Bacillus sp. TB-90 Urate Oxidase - R298C | | Descriptor: | 9-METHYL URIC ACID, HEXAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Hibi, T, Kume, A, Kawamura, A, Itoh, T, Nishiya, Y. | | Deposit date: | 2015-08-21 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Hyperstabilization of Tetrameric Bacillus sp. TB-90 Urate Oxidase by Introducing Disulfide Bonds through Structural Plasticity
Biochemistry, 55, 2016
|
|
8JVC
 
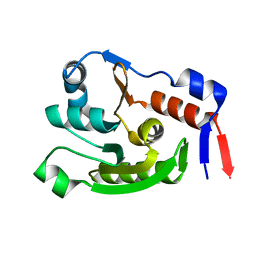 | | Crystal structure of dephospho-coenzyme A kinase | | Descriptor: | GTP-dependent dephospho-CoA kinase | | Authors: | Kita, A, Ishida, Y, Shimosaka, T, Michimori, Y, Makarova, K, Koonin, E, Atomi, H, Miki, K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of GTP-dependent dephospho-coenzyme A kinase from the hyperthermophilic archaeon, Thermococcus kodakarensis.
Proteins, 92, 2024
|
|
8JVG
 
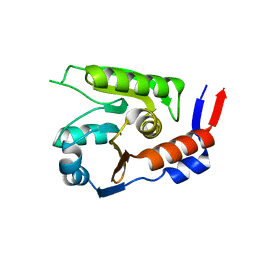 | | Crystal structure of dephospho-coenzyme A kinase | | Descriptor: | GTP-dependent dephospho-CoA kinase | | Authors: | Kita, A, Ishida, Y, Shimosaka, T, Michimori, Y, Makarova, K, Koonin, E, Atomi, H, Miki, K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of GTP-dependent dephospho-coenzyme A kinase from the hyperthermophilic archaeon, Thermococcus kodakarensis.
Proteins, 92, 2024
|
|
8JVF
 
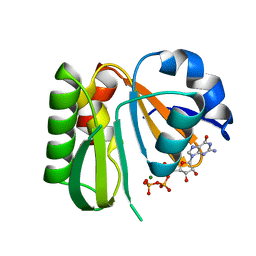 | | Crystal structure of dephospho-coenzyme A kinase | | Descriptor: | GTP-dependent dephospho-CoA kinase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Kita, A, Ishida, Y, Shimosaka, T, Michimori, Y, Makarova, K, Koonin, E, Atomi, H, Miki, K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of GTP-dependent dephospho-coenzyme A kinase from the hyperthermophilic archaeon, Thermococcus kodakarensis.
Proteins, 92, 2024
|
|
3IZ1
 
 | | C-alpha model fitted into the EM structure of Cx26M34A | | Descriptor: | Gap junction beta-2 protein | | Authors: | Oshima, A, Tani, K, Toloue, M.M, Hiroaki, Y, Smock, A, Inukai, S, Cone, A, Nicholson, B.J, Sosinsky, G.E, Fujiyoshi, Y. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (6 Å) | | Cite: | Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels.
J.Mol.Biol., 405, 2011
|
|
3IZ2
 
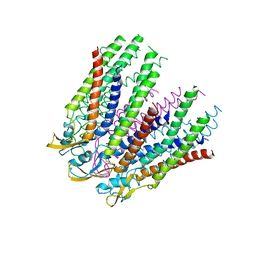 | | C-alpha model fitted into the EM structure of Cx26M34Adel2-7 | | Descriptor: | Gap junction beta-2 protein | | Authors: | Oshima, A, Tani, K, Toloue, M.M, Hiroaki, Y, Smock, A, Inukai, S, Cone, A, Nicholson, B.J, Sosinsky, G.E, Fujiyoshi, Y. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels.
J.Mol.Biol., 405, 2011
|
|
5H41
 
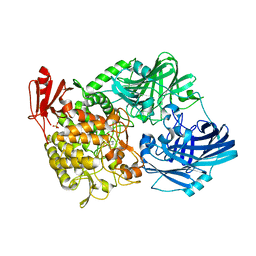 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose, isofagomine, sulfate ion | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, SULFATE ION, Uncharacterized protein, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
5H40
 
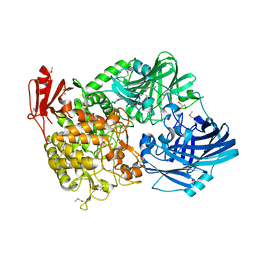 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose | | Descriptor: | CALCIUM ION, GLYCEROL, Uncharacterized protein, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
5GKI
 
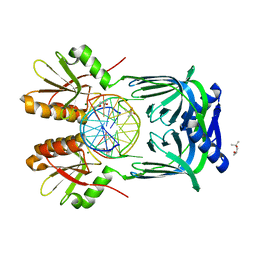 | | Structure of EndoMS-dsDNA3 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*CP*CP*TP*AP*GP*GP*TP*CP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*GP*GP*GP*CP*CP*TP*AP*GP*GP*C)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
1BQT
 
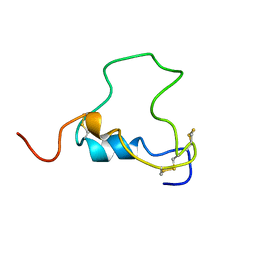 | | THREE-DIMENSIONAL STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR-I (IGF-I) DETERMINED BY 1H-NMR AND DISTANCE GEOMETRY, 6 STRUCTURES | | Descriptor: | INSULIN-LIKE GROWTH FACTOR-I | | Authors: | Sato, A, Nishimura, S, Ohkubo, T, Kyogoku, Y, Koyama, S, Kobayashi, M, Yasuda, T, Kobayashi, Y. | | Deposit date: | 1998-08-18 | | Release date: | 1999-05-18 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of human insulin-like growth factor-I (IGF-I) determined by 1H-NMR and distance geometry.
Int.J.Pept.Protein Res., 41, 1993
|
|
5GKG
 
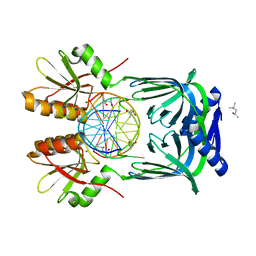 | | Structure of EndoMS-dsDNA1'' complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*CP*AP*GP*GP*TP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*CP*GP*TP*GP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKJ
 
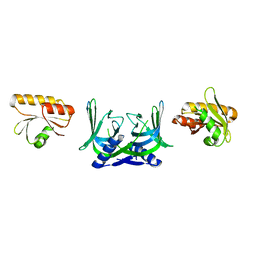 | | Structure of EndoMS in apo form | | Descriptor: | Endonuclease EndoMS | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
2Z6W
 
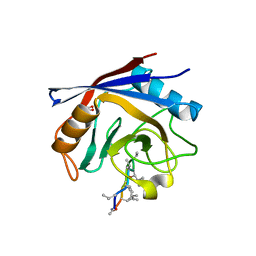 | | Crystal structure of human cyclophilin D in complex with cyclosporin A | | Descriptor: | CITRIC ACID, CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Kajitani, K, Fujihashi, M, Kobayashi, Y, Shimizu, S, Tsujimoto, Y, Miki, K. | | Deposit date: | 2007-08-09 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Crystal Structure of Human Cyclophilin D in Complex with its Inhibitor, Cyclosporin a at 0.96-A Resolution.
Proteins, 70, 2008
|
|
2YN9
 
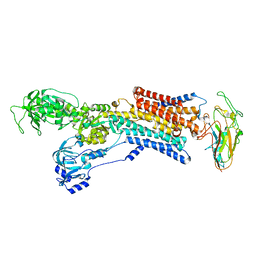 | | Cryo-EM structure of gastric H+,K+-ATPase with bound rubidium | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Friedrich, T, Fujiyoshi, Y. | | Deposit date: | 2012-10-13 | | Release date: | 2012-11-07 | | Last modified: | 2014-07-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (8 Å) | | Cite: | Cryo-Em Structure of Gastric H+,K+-ATPase with a Single Occupied Cation-Binding Site.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6KNB
 
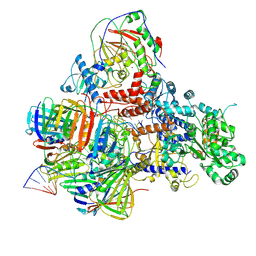 | | PolD-PCNA-DNA (form A) | | Descriptor: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | Authors: | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-05 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
2YQS
 
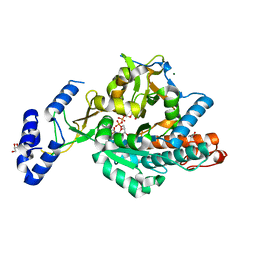 | | Crystal structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the product-binding form | | Descriptor: | GLYCEROL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
6KXE
 
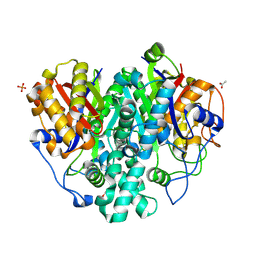 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Ketosynthase, ... | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
2YVE
 
 | | Crystal structure of the methylene blue-bound form of the multi-drug binding transcriptional repressor CgmR | | Descriptor: | 3,7-BIS(DIMETHYLAMINO)PHENOTHIAZIN-5-IUM, CHLORIDE ION, GLYCEROL, ... | | Authors: | Itou, H, Shirakihara, Y, Tanaka, I. | | Deposit date: | 2007-04-12 | | Release date: | 2008-04-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of the Multidrug Binding Repressor Corynebacteriumglutamicum CgmR in Complex with Inducers and with an Operator
J.Mol.Biol., 403, 2010
|
|
5ZSO
 
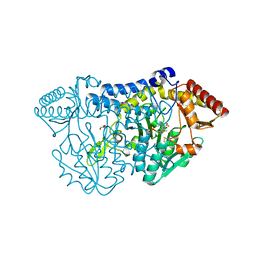 | | SufS from Bacillus subtilis, soaked with L-cysteine for 90 sec | | Descriptor: | Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Nakamura, R, Fujishiro, T, Takahashi, Y. | | Deposit date: | 2018-04-29 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 287, 2020
|
|
6KXF
 
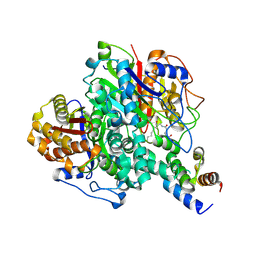 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | ACP, Ketosynthase, [(3~{R})-2,2-dimethyl-4-[[3-[2-[[(~{E})-oct-2-enoyl]amino]ethylamino]-3-oxidanylidene-propyl]amino]-3-oxidanyl-4-oxidanylidene-butyl] dihydrogen phosphate | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
2YQJ
 
 | | Crystal Structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the reaction-completed form | | Descriptor: | GLYCEROL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
2YQC
 
 | | Crystal Structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the apo-like form | | Descriptor: | GLYCEROL, MAGNESIUM ION, UDP-N-acetylglucosamine pyrophosphorylase | | Authors: | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
