6PMJ
 
 | |
6PMI
 
 | |
6O8A
 
 | | Thaumatin native-SAD structure determined at 5 keV from microcrystals | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Guo, G, Zhu, P, Fuchs, M.R, Shi, W, Andi, B, Gao, Y, Hendrickson, W.A, McSweeney, S, Liu, Q. | | 登録日 | 2019-03-09 | | 公開日 | 2019-05-08 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Synchrotron microcrystal native-SAD phasing at a low energy.
Iucrj, 6, 2019
|
|
6XQB
 
 | | SARS-CoV-2 RdRp/RNA complex | | 分子名称: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Liu, B, Shi, W, Yang, Y. | | 登録日 | 2020-07-09 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of SARS-CoV-2 RdRp/RNA complex at 3.4 Angstroms resolution
To Be Published
|
|
6XB9
 
 | | Crystal structure of Azotobacter vinelandii 3-mercaptopropionic acid dioxygenase in complex with 3-hydroxypropionic acid | | 分子名称: | 3-HYDROXY-PROPANOIC ACID, CHLORIDE ION, Cysteine dioxygenase type I protein, ... | | 著者 | Kiser, P.D, Khadka, N, Shi, W, Pierce, B.S. | | 登録日 | 2020-06-05 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure of 3-mercaptopropionic acid dioxygenase with a substrate analog reveals bidentate substrate binding at the iron center.
J.Biol.Chem., 296, 2021
|
|
6PB4
 
 | |
6PB5
 
 | |
6PBR
 
 | | Catalytic domain of E.coli dihydrolipoamide succinyltransferase in I4 space group | | 分子名称: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, SODIUM ION | | 著者 | Andi, B, Soares, A.S, Shi, W, Fuchs, M.R, McSweeney, S, Liu, Q. | | 登録日 | 2019-06-14 | | 公開日 | 2019-06-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of the dihydrolipoamide succinyltransferase catalytic domain from Escherichia coli in a novel crystal form: a tale of a common protein crystallization contaminant.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6PB6
 
 | |
6VCG
 
 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase, cobalt form | | 分子名称: | CHLORIDE ION, COBALT (II) ION, SODIUM ION, ... | | 著者 | Daruwalla, A, Shi, W, Kiser, P.D. | | 登録日 | 2019-12-20 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VCH
 
 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase in complex with 3-hydroxy-beta-apo-14'-carotenal | | 分子名称: | (2E,4E,6E,8E,10E)-11-[(4R)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-5,9-dimethylundeca-2,4,6,8,10-pentaenal, CHLORIDE ION, COBALT (II) ION, ... | | 著者 | Daruwalla, A, Shi, W, Kiser, P.D. | | 登録日 | 2019-12-20 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VCF
 
 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase, iron form | | 分子名称: | BICARBONATE ION, CHLORIDE ION, FE (II) ION, ... | | 著者 | Daruwalla, A, Shi, W, Kiser, P.D. | | 登録日 | 2019-12-20 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.687 Å) | | 主引用文献 | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8ENA
 
 | | Thaumatin native-SAD structure determined at 5 keV with a helium environmet | | 分子名称: | Thaumatin-1 | | 著者 | Karasawa, A, Andi, B, Ruchs, M.R, Shi, W, McSweeney, S, Hendrickson, W.A, Liu, Q. | | 登録日 | 2022-09-29 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Multi-crystal native-SAD phasing at 5 keV with a helium environment.
Iucrj, 9, 2022
|
|
8EN9
 
 | | TehA native-SAD structure determined at 5 keV with a helium environment | | 分子名称: | CHLORIDE ION, SODIUM ION, Tellurite resistance protein TehA homolog, ... | | 著者 | Karasawa, A, Andi, B, Ruchs, M.R, Shi, W, McSweeney, S, Hendrickson, W.A, Liu, Q. | | 登録日 | 2022-09-29 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Multi-crystal native-SAD phasing at 5 keV with a helium environment.
Iucrj, 9, 2022
|
|
8F2E
 
 | |
8FDW
 
 | | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2, ... | | 著者 | Zhang, J, Shi, W, Cai, Y.F, Zhu, H.S, Peng, H.Q, Voyer, J, Volloch, S.R, Cao, H, Mayer, M.L, Song, K.K, Xu, C, Lu, J.M, Chen, B. | | 登録日 | 2022-12-05 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-07-26 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane.
Nature, 619, 2023
|
|
1INQ
 
 | | Structure of Minor Histocompatibility Antigen peptide, H13a, complexed to H2-Db | | 分子名称: | BETA-2 MICROGLOBULIN, DIMETHYL SULFOXIDE, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | 著者 | Ostrov, D.A, Roden, M.M, Shi, W, Palmieri, E, Christianson, G.J, Mendoza, L, Villaflor, G, Tilley, D, Shastri, N, Grey, H, Almo, S.C, Roopenian, D, Nathenson, S.G. | | 登録日 | 2001-05-14 | | 公開日 | 2002-03-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | How H13 histocompatibility peptides differing by a single methyl group and lacking conventional MHC binding anchor motifs determine self-nonself discrimination.
J.Immunol., 168, 2002
|
|
4P9U
 
 | |
1JUF
 
 | | Structure of Minor Histocompatibility Antigen peptide, H13b, complexed to H2-Db | | 分子名称: | Beta-2-microglobulin, H13b peptide, H2-Db major histocompatibility antigen | | 著者 | Ostrov, D.A, Roden, M.M, Shi, W, Palmieri, E, Christianson, G.J, Mendoza, L, Villaflor, G, Tilley, D, Shastri, N, Grey, H, Almo, S.C, Roopenian, D.C, Nathenson, S.G. | | 登録日 | 2001-08-24 | | 公開日 | 2002-03-20 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | How H13 histocompatibility peptides differing by a single methyl group and lacking conventional MHC binding anchor motifs determine self-nonself discrimination.
J.Immunol., 168, 2002
|
|
6N20
 
 | |
6NCI
 
 | | Crystal structure of CDP-Chase: Vector data collection | | 分子名称: | D-ribose, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | 著者 | Miller, M.S, Shi, W, Gabelli, S.B. | | 登録日 | 2018-12-11 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6NCH
 
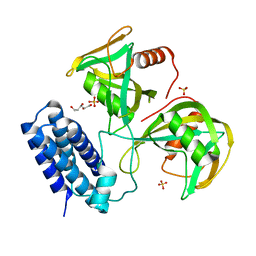 | | Crystal structure of CDP-Chase: Raster data collection | | 分子名称: | D-ribose, PHOSPHATE ION, Phosphohydrolase (MutT/nudix family protein), ... | | 著者 | Miller, M.S, Shi, W, Gabelli, S.B. | | 登録日 | 2018-12-11 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
4O63
 
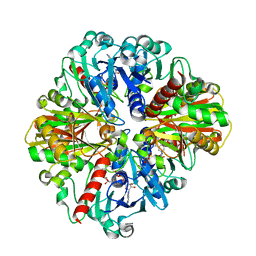 | | Co-enzyme Induced Conformational Changes in Bovine Eye Glyceraldehyde 3-Phosphate Dehydrogenase | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Baker, B.Y, Shi, W, Wang, B, Palczewski, K. | | 登録日 | 2013-12-20 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules.
Protein Sci., 23, 2014
|
|
4O59
 
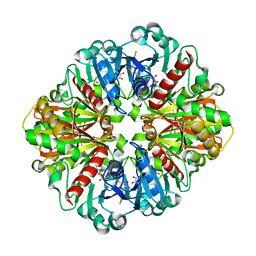 | | Co-enzyme Induced Conformational Changes in Bovine Eye Glyceraldehyde 3-Phosphate Dehydrogenase | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Baker, B.Y, Shi, W, Wang, B, Palczewski, K. | | 登録日 | 2013-12-19 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules.
Protein Sci., 23, 2014
|
|
4OU8
 
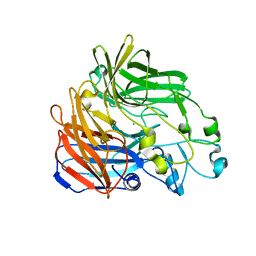 | | Crystal structure of apocarotenoid oxygenase in the presence of C8E6 | | 分子名称: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, FE (II) ION | | 著者 | Sui, X, Shi, W, Palczewski, K, Kiser, P.D. | | 登録日 | 2014-02-15 | | 公開日 | 2014-03-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Analysis of Carotenoid Isomerase Activity in a Prototypical Carotenoid Cleavage Enzyme, Apocarotenoid Oxygenase (ACO).
J.Biol.Chem., 289, 2014
|
|
