3ABZ
 
 | | Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus | | Descriptor: | Beta-glucosidase I, GLYCEROL | | Authors: | Yoshida, E, Hidaka, M, Fushinobu, S, Katayama, T, Kumagai, H. | | Deposit date: | 2009-12-25 | | Release date: | 2010-08-11 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 beta-glucosidase from Kluyveromyces marxianus.
Biochem.J., 431, 2010
|
|
3AHC
 
 | | Resting form of Phosphoketolase from Bifidobacterium Breve | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NONAETHYLENE GLYCOL, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
2B0J
 
 | | The crystal structure of the apoenzyme of the iron-sulfur-cluster-free hydrogenase (Hmd) | | Descriptor: | 5,10-methenyltetrahydromethanopterin hydrogenase | | Authors: | Pilak, O, Mamat, B, Vogt, S, Hagemeier, C.H, Thauer, R.K, Shima, S, Vonrhein, C, Warkentin, E, Ermler, U. | | Deposit date: | 2005-09-14 | | Release date: | 2006-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of the Apoenzyme of the Iron-Sulphur Cluster-free Hydrogenase
J.Mol.Biol., 358, 2006
|
|
1Z69
 
 | | Crystal structure of methylenetetrahydromethanopterin reductase (Mer) in complex with coenzyme F420 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CHLORIDE ION, COENZYME F420, ... | | Authors: | Aufhammer, S.W, Warkentin, E, Ermler, U, Hagemeier, C.H, Thauer, R.K, Shima, S. | | Deposit date: | 2005-03-22 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of methylenetetrahydromethanopterin reductase (Mer) in complex with coenzyme F420: Architecture of the F420/FMN binding site of enzymes within the nonprolyl cis-peptide containing bacterial luciferase family
Protein Sci., 14, 2005
|
|
2YYH
 
 | |
2ZQZ
 
 | | R-state structure of allosteric L-lactate dehydrogenase from Lactobacillus casei | | Descriptor: | L-lactate dehydrogenase, SULFATE ION | | Authors: | Arai, K, Ishimitsu, T, Fushinobu, S, Uchikoba, H, Matsuzawa, H, Taguchi, H. | | Deposit date: | 2008-08-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active and inactive state structures of unliganded Lactobacillus casei allosteric L-lactate dehydrogenase.
Proteins, 78, 2010
|
|
2ZSX
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 140 K: Laser on [600 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2BB9
 
 | | Structure of HIV1 protease and AKC4p_133a complex. | | Descriptor: | 2-ETHOXYETHYL (1S,2S)-3-{(2S)-4-[(3AS,8S,8AR)-2-OXO-3,3A,8,8A-TETRAHYDRO-2H-INDENO[1,2-D][1,3]OXAZOL-8-YL]-2-BENZYL-3-OXO-2,3-DIHYDRO-1H-PYRROL-2-YL}-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Smith III, A.B, Charnley, A.K, Kuo, L.C, Munshi, S. | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design, synthesis, and biological evaluation of monopyrrolinone-based HIV-1 protease inhibitors possessing augmented P2' side chains
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2D0D
 
 | | Crystal Structure of a Meta-cleavage Product Hydrolase (CumD) A129V Mutant | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, CHLORIDE ION, PHOSPHATE ION | | Authors: | Jun, S.Y, Fushinobu, S, Nojiri, H, Omori, T, Shoun, H, Wakagi, T. | | Deposit date: | 2005-08-01 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Improving the catalytic efficiency of a meta-cleavage product hydrolase (CumD) from Pseudomonas fluorescens IP01
Biochim.Biophys.Acta, 1764, 2006
|
|
3A4W
 
 | | Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, MAGNESIUM ION, ... | | Authors: | Tsuji, H, Nishimura, S, Inui, T, Ishikawa, K, Nakamura, T, Uegaki, K. | | Deposit date: | 2009-07-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and crystallographic analyses of the catalytic domain of chitinase from Pyrococcus furiosus- the role of conserved residues in the active site
Febs J., 277, 2010
|
|
2ZUW
 
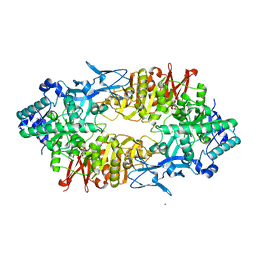 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc and sulfate | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2ZSP
 
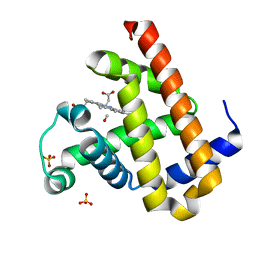 | | Carbonmonoxy Sperm Whale Myoglobin at 120 K: Laser on [300 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZSQ
 
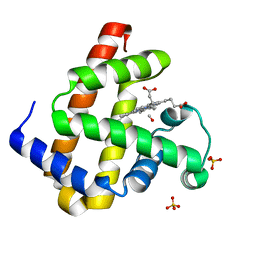 | | Carbonmonoxy Sperm Whale Myoglobin at 140 K: Laser on [150 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZSS
 
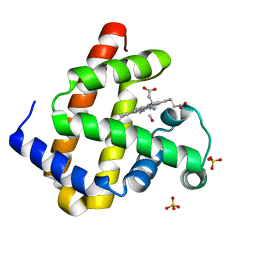 | | Carbonmonoxy Sperm Whale Myoglobin at 140 K: Laser on [300 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZT3
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 120 K: Laser on [750 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2Z1Y
 
 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase (complexed with N-(5'-phosphopyridoxyl)-L-leucine), from Thermus thermophilus HB27 | | Descriptor: | Alpha-aminodipate aminotransferase, LEUCINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2007-05-16 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of broad substrate specificity of alpha-aminoadipate aminotransferase from Thermus thermophilus
To be Published
|
|
2Z1A
 
 | | Crystal structure of 5'-nucleotidase precursor from Thermus thermophilus HB8 | | Descriptor: | 5'-nucleotidase, PHOSPHATE ION, THYMIDINE, ... | | Authors: | Nakagawa, N, Kishishita, S, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of 5'-nucleotidase precursor from Thermus thermophilus HB8
To be Published
|
|
2ZSY
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 140 K: Laser on [750 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZUT
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, GLYCEROL, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2ZSR
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 120 K: Laser on [450 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1ZC5
 
 | | Structure of the RNA signal essential for translational frameshifting in HIV-1 | | Descriptor: | HIV-1 frameshift RNA signal | | Authors: | Gaudin, C, Mazauric, M.H, Traikia, M, Guittet, E, Yoshizawa, S, Fourmy, D. | | Deposit date: | 2005-04-11 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the RNA Signal Essential for Translational Frameshifting in HIV-1
J.Mol.Biol., 349, 2005
|
|
2ZQY
 
 | | T-state structure of allosteric L-lactate dehydrogenase from Lactobacillus casei | | Descriptor: | L-lactate dehydrogenase, NITRATE ION | | Authors: | Arai, K, Ishimitsu, T, Fushinobu, S, Uchikoba, H, Matsuzawa, H, Taguchi, H. | | Deposit date: | 2008-08-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active and inactive state structures of unliganded Lactobacillus casei allosteric L-lactate dehydrogenase.
Proteins, 78, 2010
|
|
2ZUS
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase | | Descriptor: | Lacto-N-biose phosphorylase, MAGNESIUM ION | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2D4Y
 
 | |
3AO9
 
 | | Crystal structure of the C-terminal domain of sequence-specific ribonuclease | | Descriptor: | CADMIUM ION, Colicin-E5 | | Authors: | Inoue, S, Fushinobu, S, Ogawa, T, Hidaka, M, Masaki, H, Yajima, S. | | Deposit date: | 2010-09-22 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of the catalytic residues of sequence-specific and histidine-free ribonuclease colicin E5
J.Biochem., 152, 2012
|
|
