6KMF
 
 | | FimA type V pilus from P.gingivalis | | Descriptor: | Major fimbrium subunit FimA type-1 | | Authors: | Shibata, S, Shoji, M, Matsunami, H, Matthews, M, Imada, K, Nakayama, K, Wolf, M. | | Deposit date: | 2019-07-31 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
6KK9
 
 | | A Crystal structure of OspA mutant | | Descriptor: | Outer Surface Protein A | | Authors: | Shiga, S, Makabe, K. | | Deposit date: | 2019-07-24 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the beta-sheet edge of peptide self-assembly using a model protein.
Proteins, 89, 2021
|
|
7P4L
 
 | | Crystal structure of the trimeric ectodomain of archaeal Fusexin1 (Fsx1) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Fusexin1, ... | | Authors: | Nishio, S, Tunyasuvunakool, K, Jumper, J, De Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of archaeal fusexins homologous to eukaryotic HAP2/GCS1 gamete fusion proteins.
Nat Commun, 13, 2022
|
|
8J3S
 
 | | Complex structure of human cytomegalovirus protease and a macrocyclic peptide ligand | | Descriptor: | Assemblin, PHE-ILE-THR-GLY-HIS-TYR-TRP-VAL-ARG-PHE-LEU-PRO-CYS-GLY | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
8J3T
 
 | | Complex structure of human cytomegalovirus protease and a non-covalent small-molecule ligand | | Descriptor: | (4R)-1-[1-[(S)-[1-cyclopentyl-3-(2-methylphenyl)pyrazol-4-yl]-(4-methylphenyl)methyl]-2-oxidanylidene-pyridin-3-yl]-3-methyl-2-oxidanylidene-N-(3-oxidanylidene-2-azabicyclo[2.2.2]octan-4-yl)imidazolidine-4-carboxamide, Assemblin | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
1DDZ
 
 | | X-RAY STRUCTURE OF A BETA-CARBONIC ANHYDRASE FROM THE RED ALGA, PORPHYRIDIUM PURPUREUM R-1 | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Mitsuhashi, S, Mizushima, T, Yamashita, E, Miyachi, S, Tsukihara, T. | | Deposit date: | 1999-11-12 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of beta-carbonic anhydrase from the red alga, Porphyridium purpureum, reveals a novel catalytic site for CO(2) hydration.
J.Biol.Chem., 275, 2000
|
|
3LLH
 
 | | Crystal structure of the first dsRBD of TAR RNA-binding protein 2 | | Descriptor: | MALONATE ION, RISC-loading complex subunit TARBP2 | | Authors: | Yamashita, S, Kawazoe, M, Takemoto, C, Sekine, S, Wakiyama, M, Yokoyama, S. | | Deposit date: | 2010-01-29 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The structures of dsRBDs of human TRBP
To be Published
|
|
5I4F
 
 | | scFv 2D10 complexed with alpha 1,6 mannobiose | | Descriptor: | alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, scFv 2D10 | | Authors: | Vashisht, S, Kumar, A, Kaur, K.J, Salunke, D.M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Antibodies Can Exploit Molecular Crowding to Bind New Antigens at Noncanonical Paratope Positions
CHEMISTRYSELECT, 1, 2016
|
|
7NRO
 
 | | Crystal structure of AlkB in complex with manganese and N-(4-((6-((carboxymethyl)carbamoyl)-5-hydroxypyridin-2-yl)amino)phenyl)-N-oxohydroxylammonium | | Descriptor: | 2-[[6-[(4-nitrophenyl)amino]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Alpha-ketoglutarate-dependent dioxygenase AlkB, MANGANESE (II) ION | | Authors: | Shishodia, S, Maheswaran, P, Leissing, T, Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2021-03-04 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure-Based Design of Selective Fat Mass and Obesity Associated Protein (FTO) Inhibitors.
J.Med.Chem., 64, 2021
|
|
3NJU
 
 | | Crystal structure of the complex of group I phospholipase A2 with 4-Methoxy-benzoicacid at 1.4A resolution | | Descriptor: | 4-METHOXYBENZOIC ACID, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Kaushik, S, Prem Kumar, R, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the complex of group I phospholipase A2 with 4-Methoxy-benzoicacid at 1.4A resolution
To be Published
|
|
4YSI
 
 | | Structure of USP7 with a novel viral protein | | Descriptor: | GLYCEROL, SER-PRO-GLY-GLU-GLY-PRO-SER-GLY, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Chavoshi, S, Saridakis, V. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure of USP7 with a novel viral protein
J.Biol.Chem., 2016
|
|
1PMS
 
 | | PLECKSTRIN HOMOLOGY DOMAIN OF SON OF SEVENLESS 1 (SOS1) WITH GLYCINE-SERINE ADDED TO THE N-TERMINUS, NMR, 20 STRUCTURES | | Descriptor: | SOS 1 | | Authors: | Koshiba, S, Kigawa, T, Kim, J, Shirouzu, M, Bowtell, D, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-02-18 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the pleckstrin homology domain of mouse Son-of-sevenless 1 (mSos1).
J.Mol.Biol., 269, 1997
|
|
5GUX
 
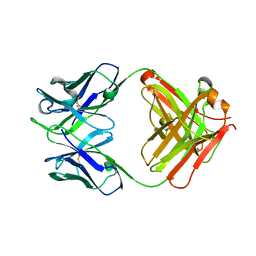 | | Cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with xenon | | Descriptor: | Antibody fab fragment heavy chain, Antibody fab fragment light chain, CALCIUM ION, ... | | Authors: | Ishii, S, Terasaka, E, Sugimoto, H, Shiro, Y, Tosha, T. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamics of nitric oxide controlled by protein complex in bacterial system.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7VNW
 
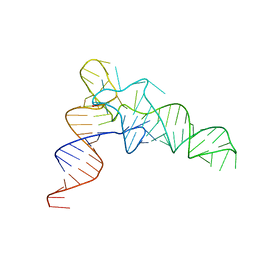 | |
7VNV
 
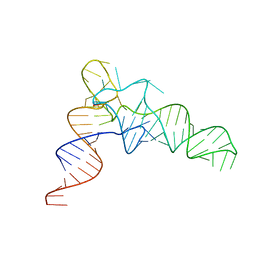 | |
7VNX
 
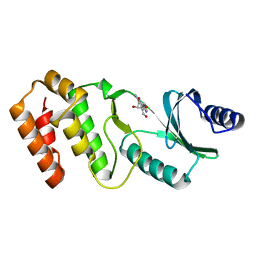 | | Crystal structure of TkArkI | | Descriptor: | GUANOSINE, TkArkI | | Authors: | Yamashita, S, Minowa, K, Ohira, T, Suzuki, T, Tomita, K. | | Deposit date: | 2021-10-12 | | Release date: | 2022-05-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Reversible RNA phosphorylation stabilizes tRNA for cellular thermotolerance.
Nature, 605, 2022
|
|
8I2Z
 
 | | Cryo-EM structure of the zeaxanthin-bound kin4B8 | | Descriptor: | RETINAL, Xanthorhodopsin, Zeaxanthin | | Authors: | Murakoshi, S, Chazan, A, Shihoya, W, Beja, O, Nureki, O. | | Deposit date: | 2023-01-15 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Phototrophy by antenna-containing rhodopsin pumps in aquatic environments.
Nature, 615, 2023
|
|
5HC9
 
 | |
5Y0P
 
 | |
5Y0S
 
 | |
5Y0R
 
 | |
5Y0N
 
 | |
8HR3
 
 | | [D-Cys5,Asp7,Val8,D-Lys16]-STp(5-17) | | Descriptor: | DCY-CYS-ASP-VAL-CYS-CYS-ASN-PRO-ALA-CYS-ALA-DLY-CYS | | Authors: | Shimamoto, S, Hidaka, Y, Yoshino, S, Goto, M. | | Deposit date: | 2022-12-14 | | Release date: | 2023-09-20 | | Method: | SOLUTION NMR | | Cite: | The Molecular Basis of Heat-Stable Enterotoxin for Vaccine Development and Cancer Cell Detection.
Molecules, 28, 2023
|
|
5Y0Q
 
 | | Crystal structure of BsTmcAL bound with AMPCPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, UPF0348 protein B4417_3650 | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis.
Nat. Chem. Biol., 14, 2018
|
|
8HR4
 
 | |
