6IDR
 
 | | Crystal structure of Vibrio cholerae MATE transporter VcmN in the bent form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
5GHK
 
 | | Crystal Structure Analysis of Canine serum albumin | | Descriptor: | Serum albumin | | Authors: | Kihira, K, Yamada, K, Kureishi, M, Yokomaku, K, Shinohara, R, Akiyama, M, Komatsu, T. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Artificial Blood for Dogs
Sci Rep, 6, 2016
|
|
1IRX
 
 | | Crystal structure of class I lysyl-tRNA synthetase | | Descriptor: | ZINC ION, lysyl-tRNA synthetase | | Authors: | Nureki, O, Terada, T, Ishitani, R, Ambrogelly, A, Ibba, M, Soll, D, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-10-25 | | Release date: | 2002-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional convergence of two lysyl-tRNA synthetases with unrelated topologies.
Nat.Struct.Biol., 9, 2002
|
|
2LHH
 
 | | Solution structure of Ca2+-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
1EIC
 
 | |
1IZR
 
 | | F46A mutant of bovine pancreatic ribonuclease A | | Descriptor: | RIBONUCLEASE A | | Authors: | Kadonosono, T, Chatani, E, Hayashi, R, Moriyama, H, Ueki, T. | | Deposit date: | 2002-10-11 | | Release date: | 2003-11-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine pancreatic RNase A
Biochemistry, 42, 2003
|
|
1IVI
 
 | | Crystal Structure of pig dihydrolipoamide dehydrogenase | | Descriptor: | dihydrolipoamide dehydrogenase | | Authors: | Toyoda, T, Kobayashi, R, Sekiguchi, T, Koike, K, Koike, M, Takenaka, A. | | Deposit date: | 2002-03-15 | | Release date: | 2003-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (8 Å) | | Cite: | Crystallization and preliminary X-ray analysis of pig E3, lipoamide dehydrogenase.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1IZP
 
 | | F46L mutant of bovine pancreatic ribonuclease A | | Descriptor: | RIBONUCLEASE A | | Authors: | Kadonosono, T, Chatani, E, Hayashi, R, Moriyama, H, Ueki, T. | | Deposit date: | 2002-10-11 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine pancreatic RNase A
Biochemistry, 42, 2003
|
|
4IKV
 
 | | Crystal structure of peptide transporter POT | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Di-tripeptide ABC transporter (Permease), OLEIC ACID, ... | | Authors: | Doki, S, Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2012-12-28 | | Release date: | 2013-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IKZ
 
 | | Crystal structure of peptide transporter POT (E310Q mutant) in complex with alafosfalin | | Descriptor: | Di-tripeptide ABC transporter (Permease), N-[(1R)-1-phosphonoethyl]-L-alaninamide, SULFATE ION | | Authors: | Doki, S, Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2012-12-28 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IKW
 
 | | Crystal structure of peptide transporter POT in complex with sulfate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Di-tripeptide ABC transporter (Permease), OLEIC ACID, ... | | Authors: | Doki, S, Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2012-12-28 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IKY
 
 | | Crystal structure of peptide transporter POT (E310Q mutant) in complex with sulfate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Di-tripeptide ABC transporter (Permease), OLEIC ACID, ... | | Authors: | Doki, S, Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2012-12-28 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4N0H
 
 | | Crystal structure of S. cerevisiae mitochondrial GatFAB | | Descriptor: | Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial, Glutamyl-tRNA(Gln) amidotransferase subunit B, ... | | Authors: | Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-10-02 | | Release date: | 2014-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases
Nucleic Acids Res., 42, 2014
|
|
3S7T
 
 | |
4N0I
 
 | | Crystal structure of S. cerevisiae mitochondrial GatFAB in complex with glutamine | | Descriptor: | GLUTAMINE, Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial, ... | | Authors: | Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-10-02 | | Release date: | 2014-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases
Nucleic Acids Res., 42, 2014
|
|
4KSD
 
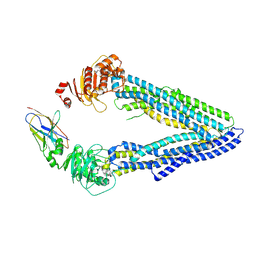 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A, R2 protein | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (4.1001 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KSC
 
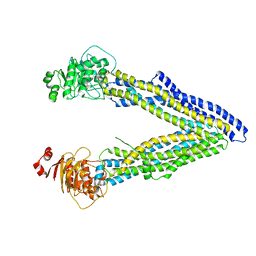 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1EIE
 
 | |
4KSB
 
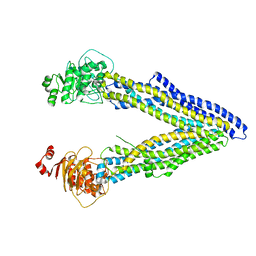 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8001 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1EKG
 
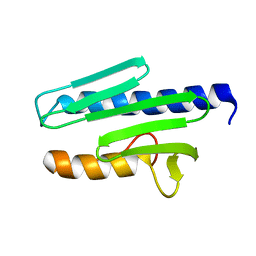 | | MATURE HUMAN FRATAXIN | | Descriptor: | FRATAXIN | | Authors: | Dhe-Paganon, S, Shigeta, R, Chi, Y.I, Ristow, M, Shoelson, S.E. | | Deposit date: | 2000-03-08 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human frataxin.
J.Biol.Chem., 275, 2000
|
|
1JSA
 
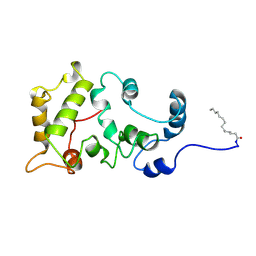 | | MYRISTOYLATED RECOVERIN WITH TWO CALCIUMS BOUND, NMR, 24 STRUCTURES | | Descriptor: | CALCIUM ION, MYRISTIC ACID, RECOVERIN | | Authors: | Ames, J.B, Ishima, R, Tanaka, T, Gordon, J.I, Stryer, L, Ikura, M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Molecular mechanics of calcium-myristoyl switches.
Nature, 389, 1997
|
|
1IZQ
 
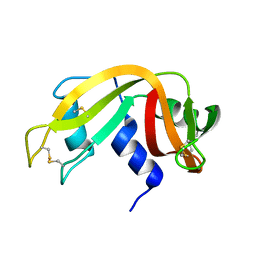 | | F46V mutant of bovine pancreatic ribonuclease A | | Descriptor: | RIBONUCLEASE A | | Authors: | Kadonosono, T, Chatani, E, Hayashi, R, Moriyama, H, Ueki, T. | | Deposit date: | 2002-10-11 | | Release date: | 2003-11-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine pancreatic RNase A
Biochemistry, 42, 2003
|
|
1FS3
 
 | |
1NT2
 
 | | CRYSTAL STRUCTURE OF FIBRILLARIN/NOP5P COMPLEX | | Descriptor: | Fibrillarin-like pre-rRNA processing protein, S-ADENOSYLMETHIONINE, conserved hypothetical protein | | Authors: | Aittaleb, M, Rashid, R, Chen, Q, Palmer, J.R, Daniels, C.J, Li, H. | | Deposit date: | 2003-01-28 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and function of archaeal box C/D sRNP core proteins.
Nat.Struct.Biol., 10, 2003
|
|
4IKX
 
 | | Crystal structure of peptide transporter POT (E310Q mutant) | | Descriptor: | Di-tripeptide ABC transporter (Permease), OLEIC ACID, SULFATE ION | | Authors: | Doki, S, Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2012-12-28 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
