6VH0
 
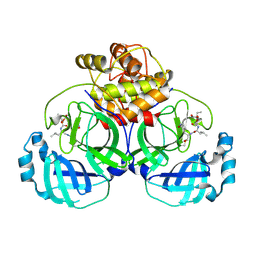 | | 1.95 A resolution structure of MERS 3CL protease in complex with inhibitor 6g | | Descriptor: | N~2~-{[(5-ethyl-1,3-dioxan-5-yl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6VH1
 
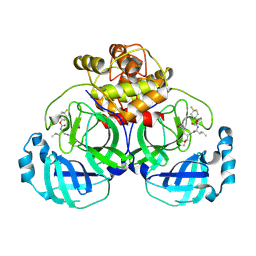 | | 2.30 A resolution structure of MERS 3CL protease in complex with inhibitor 6h | | Descriptor: | N~2~-{[(4,4-difluorocyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
2RQ4
 
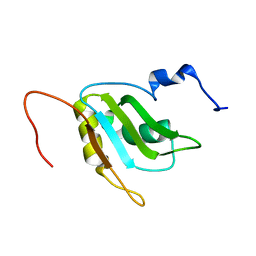 | | Refinement of RNA binding domain 3 in CUG triplet repeat RNA-binding protein 1 | | Descriptor: | CUG-BP- and ETR-3-like factor 1 | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Inoue, M, Terada, T, Kobayashi, N, Shirouzu, M, Kigawa, T, Guntert, P, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-01-19 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3
Nucleic Acids Res., 2009
|
|
6VGZ
 
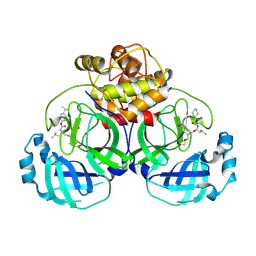 | | 2.25 A resolution structure of MERS 3CL protease in complex with inhibitor 6d | | Descriptor: | N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[trans-4-(propan-2-yl)cyclohexyl]oxy}carbonyl)-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
2RNZ
 
 | | Solution structure of the presumed chromodomain of the yeast histone acetyltransferase, Esa1 | | Descriptor: | Histone acetyltransferase ESA1 | | Authors: | Shimojo, H, Sano, N, Moriwaki, Y, Okuda, M, Horikoshi, M, Nishimura, Y. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Novel structural and functional mode of a knot essential for RNA binding activity of the Esa1 presumed chromodomain
J.Mol.Biol., 378, 2008
|
|
8SKJ
 
 | | Crystal structure of a Nanobody bound to the V5 peptide. | | Descriptor: | NbA1, V5 Epitope Tag Peptide | | Authors: | Zaghal, M, Matte, K, Venes, A, Patel, S, Laroche, G, Sarvan, S, Joshi, M, Couture, J.F, Giguere, P.M. | | Deposit date: | 2023-04-19 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Development of a V5-tag-directed nanobody and its implementation as an intracellular biosensor of GPCR signaling.
J.Biol.Chem., 299, 2023
|
|
4XF6
 
 | | myo-inositol 3-kinase bound with its products (ADP and 1D-myo-inositol 3-phosphate) | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2014-12-26 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structure and Product Analysis of an Archaeal myo-Inositol Kinase Reveal Substrate Recognition Mode and 3-OH Phosphorylation
Biochemistry, 54, 2015
|
|
2RUG
 
 | | Refined solution structure of the first RNA recognition motif domain in CPEB3 | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 3 | | Authors: | Tsuda, K, Kuwasako, K, Nagata, T, Takahashi, M, Kigawa, T, Kobayashi, N, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2014-04-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel RNA recognition motif domain in the cytoplasmic polyadenylation element binding protein 3.
Proteins, 82, 2014
|
|
4XF7
 
 | | myo-inositol 3-kinase bound with its substrates (AMPPCP and myo-inositol) | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, Carbohydrate/pyrimidine kinase, PfkB family, ... | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2014-12-26 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure and Product Analysis of an Archaeal myo-Inositol Kinase Reveal Substrate Recognition Mode and 3-OH Phosphorylation
Biochemistry, 54, 2015
|
|
6NKL
 
 | | 2.2 A resolution structure of VapBC-1 from nontypeable Haemophilus influenzae | | Descriptor: | Antitoxin VapB1, Ribonuclease VapC | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Molinaro, A.L, Daines, D.A. | | Deposit date: | 2019-01-07 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of VapBC-1 from Nontypeable Haemophilus influenzae and the Effect of PIN Domain Mutations on Survival during Infection.
J.Bacteriol., 201, 2019
|
|
6WAC
 
 | |
5B3D
 
 | | Structure of a flagellar type III secretion chaperone, FlgN | | Descriptor: | Flagella synthesis protein FlgN | | Authors: | Nakanishi, Y, Kinoshita, M, Namba, K, Minamino, T, Imada, K. | | Deposit date: | 2016-02-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rearrangements of alpha-helical structures of FlgN chaperone control the binding affinity for its cognate substrates during flagellar type III export
Mol.Microbiol., 101, 2016
|
|
2CYA
 
 | | Crystal structure of tyrosyl-tRNA synthetase from Aeropyrum pernix | | Descriptor: | SULFATE ION, Tyrosyl-tRNA synthetase | | Authors: | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Murayama, K, Chen, L, Liu, Z.J, Wang, B.C, Kuroishi, C, Kuramitsu, S, Terada, T, Bessho, Y, Shirouzu, M, Sekine, S.I, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2005
|
|
4W2I
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with negamycin, mRNA and three deacylated tRNAs in the A, P and E sites | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Szal, T, Jiang, F, Gupta, P, Matsuda, R, Shiozuka, M, Steitz, T.A, Vazquez-Laslop, N, Mankin, A.S. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Negamycin Interferes with Decoding and Translocation by Simultaneous Interaction with rRNA and tRNA.
Mol.Cell, 56, 2014
|
|
8SYI
 
 | | Cyanobacterial RNAP-EC | | Descriptor: | DNA (37-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Qayyum, M.Z, Imashimizu, M, Leanca, M, Vishwakarma, R.K, Bradley Riaz, A, Yuzenkova, Y, Murakami, K.S. | | Deposit date: | 2023-05-25 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structure and function of the Si3 insertion integrated into the trigger loop/helix of cyanobacterial RNA polymerase.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1BZA
 
 | | BETA-LACTAMASE TOHO-1 FROM ESCHERICHIA COLI TUH12191 | | Descriptor: | BETA-LACTAMASE, SULFATE ION | | Authors: | Ibuka, A, Taguchi, A, Ishiguro, M, Fushinobu, S, Ishii, Y, Kamitori, S, Okuyama, K, Yamaguchi, K, Konno, M, Matsuzawa, H. | | Deposit date: | 1998-10-28 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the E166A mutant of extended-spectrum beta-lactamase Toho-1 at 1.8 A resolution.
J.Mol.Biol., 285, 1999
|
|
3WQB
 
 | | Crystal structure of aeromonas sobria serine protease (ASP) and the chaperone (ORF2) complex | | Descriptor: | CALCIUM ION, Extracellular serine protease, Open reading frame 2 | | Authors: | Kobayashi, H, Yoshida, T, Miyakawa, T, Kato, R, Tashiro, M, Yamanaka, H, Tanokura, M, Tsuge, H. | | Deposit date: | 2014-01-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural Basis for Action of the External Chaperone for a Propeptide-deficient Serine Protease from Aeromonas sobria.
J.Biol.Chem., 290, 2015
|
|
6M7G
 
 | | Crystal structure of ArsN, N-acetyltransferase with substrate phosphinothricin from Pseudomonas putida KT2440 | | Descriptor: | PHOSPHINOTHRICIN, Phosphinothricin N-acetyltransferase | | Authors: | Venkadesh, S, Dheeman, D.S, Yoshinaga, M, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.657 Å) | | Cite: | Arsinothricin, an arsenic-containing non-proteinogenic amino acid analog of glutamate, is a broad-spectrum antibiotic.
Commun Biol, 2, 2019
|
|
4YCL
 
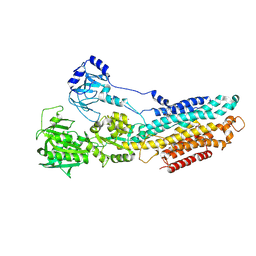 | | Crystal structure of the SR CA2+-ATPASE with bound CPA | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Ogawa, H, Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7Y4A
 
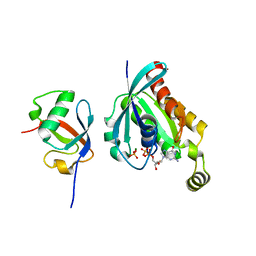 | | Crystal structure of human ELMO1 RBD-RhoG complex | | Descriptor: | Engulfment and cell motility protein 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tsuda, K, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Targeting Ras-binding domain of ELMO1 by computational nanobody design.
Commun Biol, 6, 2023
|
|
7DPA
 
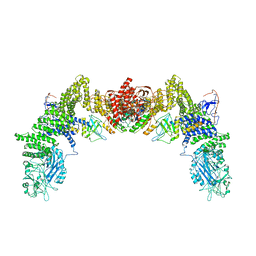 | | Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Kaushik, R, Ehara, H, Yokoyama, T, Uchikubo-Kamo, T, Mishima-Tsumagari, C, Yonemochi, M, Ikeda, M, Hanada, K, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2020-12-18 | | Release date: | 2021-08-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex.
Sci Adv, 7, 2021
|
|
7Y6L
 
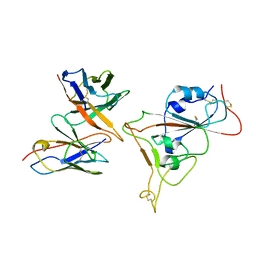 | |
7Y6N
 
 | |
1WWM
 
 | | Crystal Structure of Conserved Hypothetical Protein TT2028 from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TT2028 | | Authors: | Mizohata, E, Ushikoshi-Nakayama, R, Terada, T, Murayama, K, Sakai, H, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-07 | | Release date: | 2005-07-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of TT2028 from an Extremely Thermophilic Bacterium Thermus thermophilus HB8
To be Published
|
|
7YV1
 
 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor LUNA18 and KA30L Fab | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, KA30L Fab H-chain, ... | | Authors: | Irie, M, Fukami, T.A, Matsuo, A, Saka, K, Nishimura, M, Saito, H, Torizawa, T, Tanada, M, Ohta, A. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
J.Am.Chem.Soc., 145, 2023
|
|
