5GTQ
 
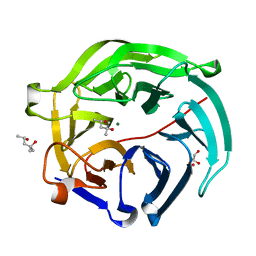 | | Luciferin-regenerating enzyme at cryogenic temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Luciferin regenerating enzyme, ... | | Authors: | Yamashita, K, Murai, T, Yamamoto, M, Gomi, K, Kajiyama, N, Kato, H, Nakatsu, T. | | Deposit date: | 2016-08-23 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Luciferin-regenerating enzyme at cryogenic temperature
To Be Published
|
|
1F3K
 
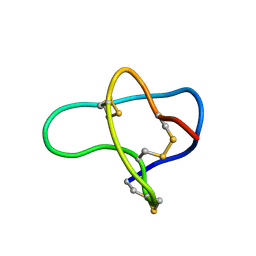 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF OMEGA-CONOTOXIN TXVII, AN L-TYPE CALCIUM CHANNEL BLOCKER | | Descriptor: | OMEGA-CONOTOXIN TXVII | | Authors: | Kobayashi, K, Sasaki, T, Sato, K, Kohno, T. | | Deposit date: | 2000-06-05 | | Release date: | 2000-12-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of omega-conotoxin TxVII, an L-type calcium channel blocker.
Biochemistry, 39, 2000
|
|
3JTH
 
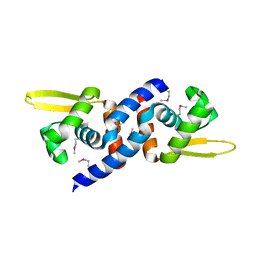 | |
8V5T
 
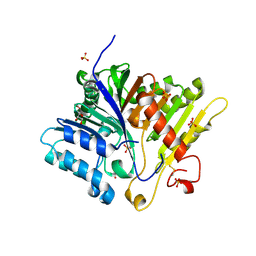 | | Crystal structure of Alzheimers disease phospholipase D3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, GLYCEROL, ... | | Authors: | Ishii, K, Hermans, S.J, Nero, T.L, Gorman, M.A, Parker, M.W. | | Deposit date: | 2023-12-01 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Alzheimer's disease phospholipase D3 provides a molecular basis for understanding its normal and pathological functions.
Febs J., 2024
|
|
3VMF
 
 | | Archaeal protein | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kobayashi, K, Saito, K, Ishitani, R, Ito, K, Nureki, O. | | Deposit date: | 2011-12-12 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for translation termination by archaeal RF1 and GTP-bound EF1alpha complex
Nucleic Acids Res., 40, 2012
|
|
8WT1
 
 | | Crystal structure of S9 carboxypeptidase from Geobacillus sterothermophilus | | Descriptor: | ALANINE, CITRATE ANION, GLYCEROL, ... | | Authors: | Chandravanshi, K, Kumar, A, Sen, C, Singh, R, Bhange, G.B, Makde, R.D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and solution scattering of Geobacillus stearothermophilus S9 peptidase reveal structural adaptations for carboxypeptidase activity.
Febs Lett., 598, 2024
|
|
3WTR
 
 | | Crystal structure of E. coli YfcM bound to Co(II) | | Descriptor: | COBALT (II) ION, Uncharacterized protein | | Authors: | Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2014-04-19 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The non-canonical hydroxylase structure of YfcM reveals a metal ion-coordination motif required for EF-P hydroxylation
Nucleic Acids Res., 42, 2014
|
|
3WXM
 
 | | Crystal structure of archaeal Pelota and GTP-bound EF1 alpha complex | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kobayashi, K, Ishitani, R, Nureki, O. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1 alpha complex
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4KXT
 
 | |
4F1N
 
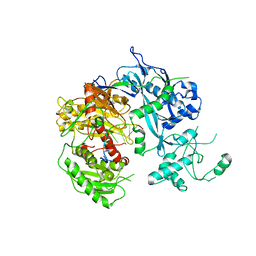 | | Crystal structure of Kluyveromyces polysporus Argonaute with a guide RNA | | Descriptor: | KpAGO, RNA 5'-R(P*UP*AP*AP*AP*AP*AP*AP*AP*A)-3' | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2012-05-07 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Structure of yeast Argonaute with guide RNA.
Nature, 486, 2012
|
|
1NYJ
 
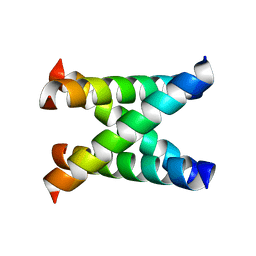 | |
4HPC
 
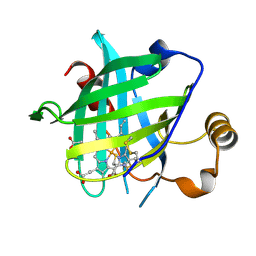 | |
4HPB
 
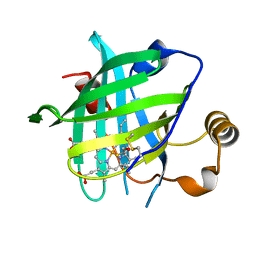 | |
4HPD
 
 | |
4HPA
 
 | |
7VLK
 
 | | eIF2B-SFSV NSs C2-imposed | | Descriptor: | Non-structural protein NS-S, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-10-04 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
7VUC
 
 | |
2AXU
 
 | | Structure of PrgX | | Descriptor: | PrgX | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.K, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-06 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AWI
 
 | | Structure of PrgX Y153C mutant | | Descriptor: | PrgX | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.k, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-01 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AW6
 
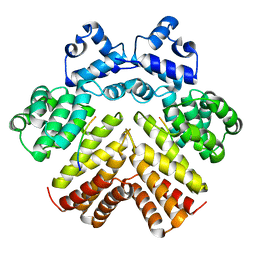 | | Structure of a bacterial peptide pheromone/receptor complex and its mechanism of gene regulation | | Descriptor: | PrgX, peptide | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.K, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-08-31 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7CGR
 
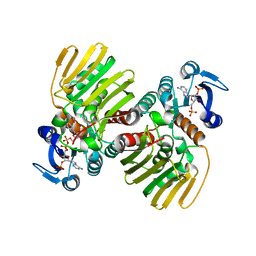 | |
6JLZ
 
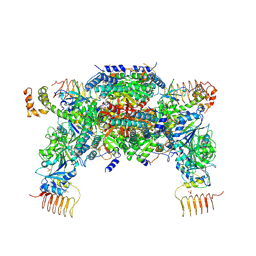 | | P-eIF2a - eIF2B complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, PHOSPHATE ION, Probable translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-01 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis for eIF2B inhibition in integrated stress response.
Science, 364, 2019
|
|
6JLY
 
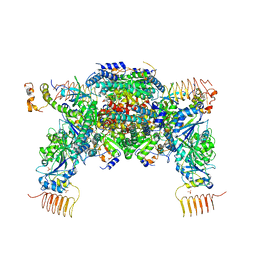 | | eIF2a - eIF2B complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, PHOSPHATE ION, Probable translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for eIF2B inhibition in integrated stress response.
Science, 364, 2019
|
|
7D45
 
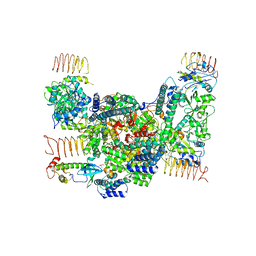 | | eIF2B-eIF2(aP), aP1 complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2020-09-22 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | ISRIB Blunts the Integrated Stress Response by Allosterically Antagonising the Inhibitory Effect of Phosphorylated eIF2 on eIF2B.
Mol.Cell, 81, 2021
|
|
7DO6
 
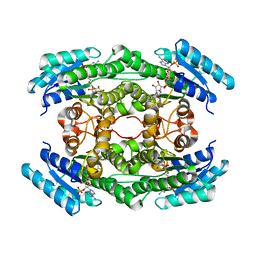 | |
