2ENW
 
 | |
2EJJ
 
 | |
2ELD
 
 | |
2E4R
 
 | |
2ED3
 
 | |
2EMU
 
 | |
2ENU
 
 | |
2EMR
 
 | |
2EN5
 
 | |
2E4N
 
 | |
2EH4
 
 | |
2ED5
 
 | |
2ELE
 
 | |
2EH5
 
 | |
2DVL
 
 | |
2DY0
 
 | |
2CSX
 
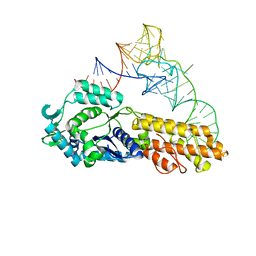 | | Crystal structure of Aquifex aeolicus methionyl-tRNA synthetase complexed with tRNA(Met) | | Descriptor: | Methionyl-tRNA synthetase, RNA (75-MER) | | Authors: | Nakanishi, K, Ogiso, Y, Nakama, T, Fukai, S, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for anticodon recognition by methionyl-tRNA synthetase.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2CT8
 
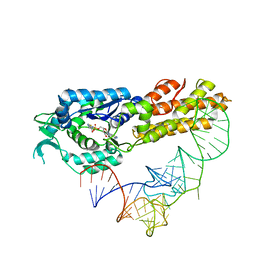 | | Crystal structure of Aquifex aeolicus methionyl-tRNA synthetase complexed with tRNA(Met) and methionyl-adenylate anologue | | Descriptor: | 5'-O-[(L-METHIONYL)-SULPHAMOYL]ADENOSINE, Methionyl-tRNA synthetase, RNA (74-MER) | | Authors: | Nakanishi, K, Ogiso, Y, Nakama, T, Fukai, S, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for anticodon recognition by methionyl-tRNA synthetase.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2E9Y
 
 | |
2ENI
 
 | |
2DQX
 
 | | mutant beta-amylase (W55R) from soy bean | | Descriptor: | Beta-amylase | | Authors: | Ishikawa, K. | | Deposit date: | 2006-06-01 | | Release date: | 2007-05-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetic and structural analysis of enzyme sliding on a substrate: multiple attack in beta-amylase
Biochemistry, 46, 2007
|
|
2GRB
 
 | |
4PT6
 
 | | The discobody: an engineered discoidin domain from factor VIII that binds v 3 integrin with antibody-like affinities | | Descriptor: | Coagulation factor VIII | | Authors: | Kym, G, Shi, K, Kamajaya, A, Hamdouche, S, Kostecki, J.S, Wannier, T, Li, B, Nikolovski, P, Mayo, S.L. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The discobody: an engineered discoidin domain from factor VIII that binds v 3 integrin with antibody-like affinities
To be Published
|
|
6E8C
 
 | | Crystal structure of the double homeodomain of DUX4 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), Double homeobox protein 4 | | Authors: | Lee, J.K, Bosnakovski, D, Toso, E.A, Dinh, T, Banerjee, S, Bohl, T.E, Shi, K, Kurahashi, K, Kyba, M, Aihara, H. | | Deposit date: | 2018-07-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of the Double Homeodomain of DUX4 in Complex with DNA.
Cell Rep, 25, 2018
|
|
5W8N
 
 | |
