3IH8
 
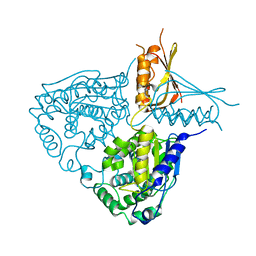 | | Crystal Structure Analysis of Mglu in its native form | | Descriptor: | Salt-tolerant glutaminase | | Authors: | Yoshimune, K, Shirakihara, Y. | | Deposit date: | 2009-07-29 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of salt-tolerant glutaminase from Micrococcus luteus K-3 in the presence and absence of its product l-glutamate and its activator Tris
Febs J., 277, 2010
|
|
3IHB
 
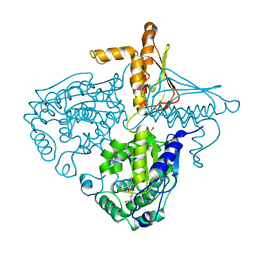 | | Crystal Structure Analysis of Mglu in its tris and glutamate form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTAMIC ACID, Salt-tolerant glutaminase | | Authors: | Yoshimune, K, Shirakihara, Y. | | Deposit date: | 2009-07-29 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of salt-tolerant glutaminase from Micrococcus luteus K-3 in the presence and absence of its product l-glutamate and its activator Tris
Febs J., 277, 2010
|
|
4DOQ
 
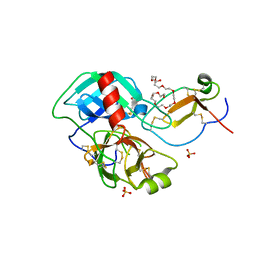 | | Crystal structure of the complex of Porcine Pancreatic Trypsin with 1/2SLPI | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Antileukoproteinase, CALCIUM ION, ... | | Authors: | Fukushima, K, Takimoto-Kamimura, M. | | Deposit date: | 2012-02-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure basis 1/2SLPI and porcine pancreas trypsin interaction
J.SYNCHROTRON RADIAT., 20, 2013
|
|
4U6O
 
 | |
8IMR
 
 | | Structure of ligand-free human macrophage migration inhibitory factor | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Sugishima, K, Noguchi, K, Yohda, M, Odaka, M, Matsumura, H. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification of methotrexate as an inhibitor of macrophage migration inhibitory factor by high-resolution crystal structure analysis
To Be Published
|
|
8GW8
 
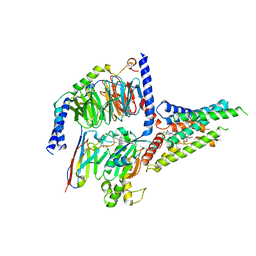 | | the human PTH1 receptor bound to an intracellular biased agonist | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform Gnas-2 of Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Okamoto, H.H, Nureki, O. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Class B1 GPCR activation by an intracellular agonist.
Nature, 618, 2023
|
|
4YVH
 
 | |
4YVI
 
 | |
4YVJ
 
 | |
4YVK
 
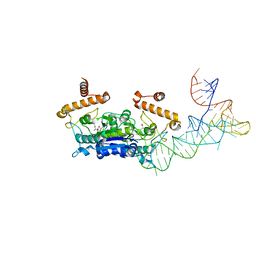 | |
5B5K
 
 | | Crystal structure of Izumo1, the mammalian sperm ligand for egg Juno | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Izumo sperm-egg fusion protein 1 | | Authors: | Nishimura, K, Han, L, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-05-11 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of sperm Izumo1 reveals unexpected similarities with Plasmodium invasion proteins.
Curr.Biol., 26, 2016
|
|
4PDN
 
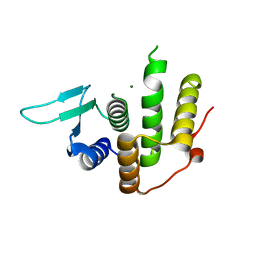 | | Crystal structure of E. coli YfcM | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2014-04-19 | | Release date: | 2015-03-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | The non-canonical hydroxylase structure of YfcM reveals a metal ion-coordination motif required for EF-P hydroxylation.
Nucleic Acids Res., 42, 2014
|
|
8V7O
 
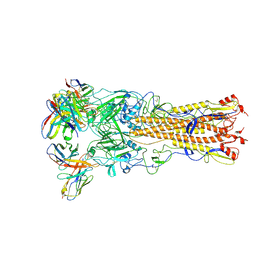 | |
6LU4
 
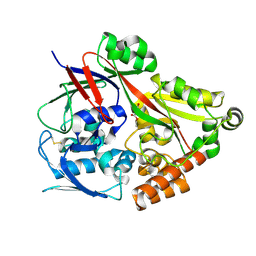 | | Crystal structure of the substrate binding protein from Microbacterium hydrocarbonoxydans complexed with propylparaben | | Descriptor: | Substrate binding protein, propyl 4-hydroxybenzoate | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
3WXM
 
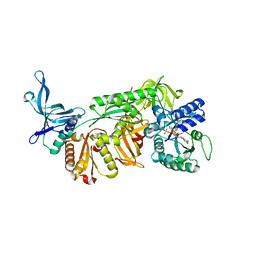 | | Crystal structure of archaeal Pelota and GTP-bound EF1 alpha complex | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kobayashi, K, Ishitani, R, Nureki, O. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1 alpha complex
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3VMF
 
 | | Archaeal protein | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kobayashi, K, Saito, K, Ishitani, R, Ito, K, Nureki, O. | | Deposit date: | 2011-12-12 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for translation termination by archaeal RF1 and GTP-bound EF1alpha complex
Nucleic Acids Res., 40, 2012
|
|
7EHZ
 
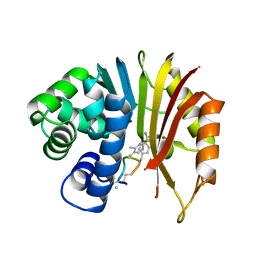 | | Structure of human NNMT in complex with macrocyclic peptide 2 | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide 2 | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-30 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
7EGU
 
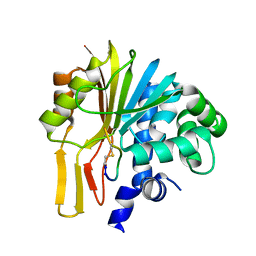 | | Structure of human NNMT in complex with macrocyclic peptide X | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide X | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-26 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
5BUP
 
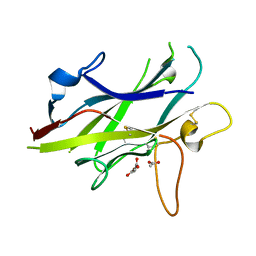 | | Crystal structure of the ZP-C domain of mouse ZP2 | | Descriptor: | ACETATE ION, Zona pellucida sperm-binding protein 2 | | Authors: | Nishimura, K, Jovine, L. | | Deposit date: | 2015-06-04 | | Release date: | 2016-01-27 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | A structured interdomain linker directs self-polymerization of human uromodulin.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7EI2
 
 | | Structure of human NNMT in complex with macrocyclic peptide 8 | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide 8 | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-30 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
6MTI
 
 | | Synaptotagmin-1 C2A, C2B domains and SNARE-pin proteins (5CCI) individually docked into Cryo-EM map of C2AB-SNARE complexes helically organized on lipid nanotube surface in presence of Mg2+ | | Descriptor: | MAGNESIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Grushin, K, Wang, J, Coleman, J, Rothman, J, Sindelar, C, Krishnakumar, S. | | Deposit date: | 2018-10-19 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural basis for the clamping and Ca2+activation of SNARE-mediated fusion by synaptotagmin.
Nat Commun, 10, 2019
|
|
4F1N
 
 | | Crystal structure of Kluyveromyces polysporus Argonaute with a guide RNA | | Descriptor: | KpAGO, RNA 5'-R(P*UP*AP*AP*AP*AP*AP*AP*AP*A)-3' | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2012-05-07 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Structure of yeast Argonaute with guide RNA.
Nature, 486, 2012
|
|
2ENY
 
 | | Solution structure of the ig-like domain (2735-2825) of human obscurin | | Descriptor: | Obscurin | | Authors: | Nagashima, K, Izumi, K, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ig-like domain (2735-2825) of human obscurin
To be Published
|
|
1DET
 
 | | RIBONUCLEASE T1 CARBOXYMETHYLATED AT GLU 58 IN COMPLEX WITH 2'GMP | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Ishikawa, K, Suzuki, E, Tanokura, M, Takahashi, K. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of ribonuclease T1 carboxymethylated at Glu58 in complex with 2'-GMP.
Biochemistry, 35, 1996
|
|
1IW8
 
 | | Crystal Structure of a mutant of acid phosphatase from Escherichia blattae (G74D/I153T) | | Descriptor: | SULFATE ION, acid phosphatase | | Authors: | Ishikawa, K, Mihara, Y, Shimba, N, Ohtsu, N, Kawasaki, H, Suzuki, E, Asano, Y. | | Deposit date: | 2002-04-22 | | Release date: | 2002-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enhancement of nucleoside phosphorylation activity in an acid phosphatase
PROTEIN ENG., 15, 2002
|
|
