2ZPH
 
 | | Complex of Fe-type nitrile hydratase with tert-butylisonitrile, photo-activated for 340min at 293K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
5YZ7
 
 | | Crystal structure of OsD14 in complex with D-ring-opened 7'-carba-4BD | | Descriptor: | (2Z,4S)-5-(4-bromophenyl)-4-hydroxy-2-methylpent-2-enoic acid, Strigolactone esterase D14 | | Authors: | Hirabayashi, K, Jiang, K, Xu, Y, Miyakawa, T, Asami, T, Tanokura, M. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Rationally Designed Strigolactone Analogs as Antagonists of the D14 Receptor.
Plant Cell Physiol., 59, 2018
|
|
5XLH
 
 | | Crystal structure of aerobically purified and aerobically crystallized for 12weeks D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
1V8G
 
 | |
2ZPB
 
 | | nitrosylated Fe-type nitrile hydratase | | Descriptor: | FE (III) ION, MAGNESIUM ION, NITRIC OXIDE, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-09 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
1VGO
 
 | | Crystal Structure of Archaerhodopsin-2 | | Descriptor: | Archaerhodopsin 2, RETINAL, SULFATE ION, ... | | Authors: | Yoshimura, K, Enami, N, Murakami, M, Okumura, H, Ihara, K, Kouyama, T. | | Deposit date: | 2004-04-28 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of archaerhodopsin-1 and -2: Common structural motif in archaeal light-driven proton pumps
J.Mol.Biol., 358, 2006
|
|
1WLF
 
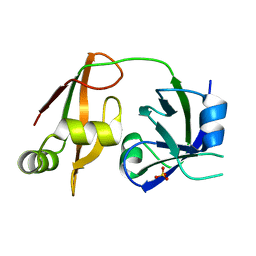 | | Structure of the N-terminal domain of PEX1 AAA-ATPase: Characterization of a putative adaptor-binding domain | | Descriptor: | Peroxisome biogenesis factor 1, SULFATE ION | | Authors: | Shiozawa, K, Maita, N, Tomii, K, Seto, A, Goda, N, Tochio, H, Akiyama, Y, Shimizu, T, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2004-06-25 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the N-terminal Domain of PEX1 AAA-ATPase: CHARACTERIZATION OF A PUTATIVE ADAPTOR-BINDING DOMAIN
J.Biol.Chem., 279, 2004
|
|
1WQ5
 
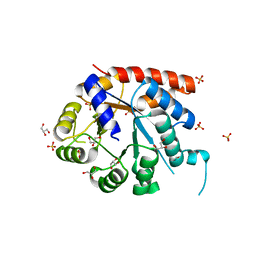 | | Crystal structure of tryptophan synthase alpha-subunit from Escherichia coli | | Descriptor: | GLYCEROL, SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Nishio, K, Morimoto, Y, Ishizuka, M, Ogasahara, K, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-09-22 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Changes in the alpha-Subunit Coupled to Binding of the beta(2)-Subunit of Tryptophan Synthase from Escherichia coli: Crystal Structure of the Tryptophan Synthase alpha-Subunit Alon
Biochemistry, 44, 2005
|
|
3B07
 
 | | Crystal structure of octameric pore form of gamma-hemolysin from Staphylococcus aureus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Gamma-hemolysin component A, Gamma-hemolysin component B | | Authors: | Yamashita, K, Kawai, Y, Tanaka, Y, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-06 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Crystal structure of the octameric pore of
staphylococcal gamma-hemolysin reveals the beta-barrel
pore formation mechanism by two components
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2ZV3
 
 | |
2D29
 
 | |
2D3K
 
 | |
2ZVC
 
 | |
2ZVB
 
 | |
3ANZ
 
 | | Crystal Structure of alpha-hemolysin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Alpha-hemolysin | | Authors: | Yamashita, K, Kawauchi, H, Tanaka, Y, Yao, M, Tanaka, I. | | Deposit date: | 2010-09-16 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | 2-Methyl-2,4-pentanediol induces spontaneous assembly of staphylococcal alpha-hemolysin into heptameric pore structure
Protein Sci., 20, 2011
|
|
1RY6
 
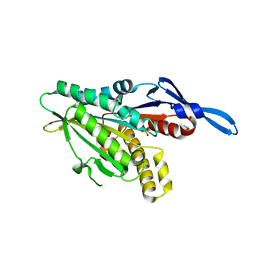 | | Crystal Structure of Internal Kinesin Motor Domain | | Descriptor: | INTERNAL KINESIN, SULFATE ION | | Authors: | Shipley, K, Hekmat-Nejad, M, Turner, J, Moores, C, Anderson, R, Milligan, R, Sakowicz, R, Fletterick, R. | | Deposit date: | 2003-12-19 | | Release date: | 2004-04-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a kinesin microtubule depolymerization machine.
Embo J., 23, 2004
|
|
1X1O
 
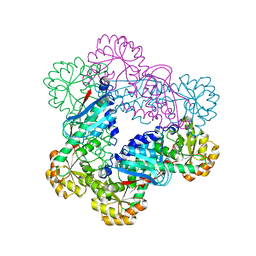 | |
1WN2
 
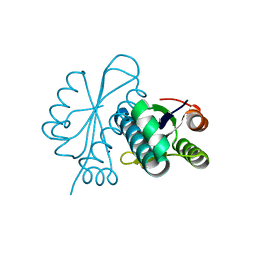 | |
1WS9
 
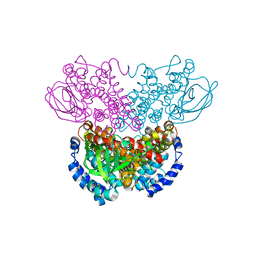 | |
1WU8
 
 | |
3AI7
 
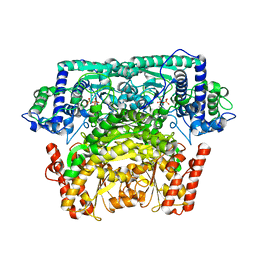 | | Crystal Structure of Bifidobacterium Longum Phosphoketolase | | Descriptor: | CALCIUM ION, THIAMINE DIPHOSPHATE, Xylulose-5-phosphate/fructose-6-phosphate phosphoketolase | | Authors: | Takahashi, K, Tagami, U, Shimba, N, Kashiwagi, T, Ishikawa, K, Suzuki, E. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Bifidobacterium Longum phosphoketolase; key enzyme for glucose metabolism in Bifidobacterium
Febs Lett., 584, 2010
|
|
2ZTM
 
 | | T190S mutant of D-3-hydroxybutyrate dehydrogenase | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-07 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
3A7S
 
 | | Catalytic domain of UCH37 | | Descriptor: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Nishio, K, Kim, S.W, Kawai, K, Mizushima, T, Yamane, T, Hamazaki, J, Murata, S, Tanaka, K. | | Deposit date: | 2009-10-04 | | Release date: | 2009-11-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the de-ubiquitinating enzyme UCH37 (human UCH-L5) catalytic domain
Biochem.Biophys.Res.Commun., 2009
|
|
2ZTL
 
 | | Closed conformation of D-3-hydroxybutyrate dehydrogenase complexed with NAD+ and L-3-hydroxybutyrate | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, D(-)-3-hydroxybutyrate dehydrogenase, GLYCEROL, ... | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-07 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
2ZTU
 
 | | T190A mutant of D-3-hydroxybutyrate dehydrogenase complexed with NAD+ | | Descriptor: | D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Nakashima, K, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-10-09 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Closed complex of the D-3-hydroxybutyrate dehydrogenase induced by an enantiomeric competitive inhibitor.
J.Biochem., 145, 2009
|
|
