3W6N
 
 | | Crystal structure of human Dlp1 in complex with GMP-PN.Pi | | Descriptor: | AMINOPHOSPHONIC ACID-GUANYLATE ESTER, CALCIUM ION, Dynamin-1-like protein, ... | | Authors: | Kishida, H, Sugio, S. | | Deposit date: | 2013-02-17 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GTPase domain fused with minimal stalks from human dynamin-1-like protein (Dlp1) in complex with several nucleotide analogues
CURR TOP PEPT PROTEIN RES., 14, 2013
|
|
3W6P
 
 | | Crystal structure of human Dlp1 in complex with GDP.AlF4 | | Descriptor: | CALCIUM ION, Dynamin-1-like protein, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kishida, H, Sugio, S. | | Deposit date: | 2013-02-17 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of GTPase domain fused with minimal stalks from human dynamin-1-like protein (Dlp1) in complex with several nucleotide analogues
CURR TOP PEPT PROTEIN RES., 14, 2013
|
|
3W6O
 
 | | Crystal structure of human Dlp1 in complex with GMP-PCP | | Descriptor: | CALCIUM ION, Dynamin-1-like protein, MAGNESIUM ION, ... | | Authors: | Kishida, H, Sugio, S. | | Deposit date: | 2013-02-17 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of GTPase domain fused with minimal stalks from human dynamin-1-like protein (Dlp1) in complex with several nucleotide analogues
CURR TOP PEPT PROTEIN RES., 14, 2013
|
|
3WV6
 
 | |
2E2P
 
 | | Crystal structure of Sulfolobus tokodaii hexokinase in complex with ADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE, ... | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
2E2Q
 
 | | Crystal structure of Sulfolobus tokodaii hexokinase in complex with xylose, Mg2+, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE, MAGNESIUM ION, ... | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
2E2N
 
 | | Crystal structure of Sulfolobus tokodaii hexokinase in the apo form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEXOKINASE, SULFATE ION | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
2E2O
 
 | | Crystal structure of Sulfolobus tokodaii hexokinase in complex with glucose | | Descriptor: | HEXOKINASE, beta-D-glucopyranose | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
2Z5V
 
 | | Solution structure of the TIR domain of human MyD88 | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Ohnishi, H, Tochio, H, Hiroaki, H, Kondo, N, Kato, Z, Shirakawa, M. | | Deposit date: | 2007-07-19 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling.
Proc.Natl.Acad.Sci.USA, 2009
|
|
2COV
 
 | | Crystal structure of CBM31 from beta-1,3-xylanase | | Descriptor: | beta-1,3-xylanase | | Authors: | Hashimoto, H, Tamai, Y, Okazaki, F, Tamaru, Y, Shimizu, T, Araki, T, Sato, M. | | Deposit date: | 2005-05-18 | | Release date: | 2005-09-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The first crystal structure of a family 31 carbohydrate-binding module with affinity to beta-1,3-xylan
Febs Lett., 579, 2005
|
|
2CFM
 
 | |
2ELG
 
 | | The rare crystallographic structure of d(CGCGCG)2: The natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)2 at 10 degree celsius | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DG)-3'), MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ohishi, H, Tozuka, Y, Zhou, D.Y, Ishida, T, Nakatani, K. | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The rare crystallographic structure of d(CGCGCG)(2): The natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)(2) at 10 degrees C
Biochem.Biophys.Res.Commun., 358, 2007
|
|
336D
 
 | | INTERACTION BETWEEN LEFT-HANDED Z-DNA AND POLYAMINE-3 THE CRYSTAL STRUCTURE OF THE D(CG)3 AND THERMOSPERMINE COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, N-(3-AMINO-PROPYL)-N-(5-AMINOPROPYL)-1,4-DIAMINOBUTANE | | Authors: | Ohishi, H, Terasoma, N, Nakanishi, I, Van Der Marel, G, Van Boom, J.H, Rich, A, Wang, A.H.-J, Hakoshima, T, Tomita, K.-I. | | Deposit date: | 1997-06-24 | | Release date: | 1998-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Interaction between left-handed Z-DNA and polyamine - 3. The crystal structure of the d(CG)3 and thermospermine complex.
FEBS Lett., 398, 1996
|
|
2DDD
 
 | | Unique behavior of a histidine responsible for an engineered green-to-red photoconversion process | | Descriptor: | MAGNESIUM ION, SODIUM ION, photoconvertible fluorescent protein | | Authors: | Shimizu, H, Tsutsui, H, Nukina, N, Miyawaki, A. | | Deposit date: | 2006-01-27 | | Release date: | 2006-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The E1 mechanism in photo-induced beta-elimination reactions for green-to-red conversion of fluorescent proteins
Chem.Biol., 16, 2009
|
|
2DDC
 
 | | Unique behavior of a histidine responsible for an engineered green-to-red photoconversion process | | Descriptor: | MAGNESIUM ION, SODIUM ION, photoconvertible fluorescent protein | | Authors: | Shimizu, H, Tsutsui, H, Nukina, N, Miyawaki, A. | | Deposit date: | 2006-01-27 | | Release date: | 2006-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The E1 mechanism in photo-induced beta-elimination reactions for green-to-red conversion of fluorescent proteins.
Chem.Biol., 16, 2009
|
|
3A2F
 
 | |
3AX7
 
 | | Bovine Xanthine Oxidase, protease cleaved form | | Descriptor: | 2-HYDROXYBENZOIC ACID, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Ishikita, H, Eger, B.T, Pai, E.F, Okamoto, K, Nishino, T. | | Deposit date: | 2011-03-30 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase
J.Am.Chem.Soc., 134, 2012
|
|
3AX9
 
 | | Bovine xanthine oxidase, protease cleaved form | | Descriptor: | 2-HYDROXYBENZOIC ACID, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Ishikita, H, Eger, B.T, Pai, E.F, Okamoto, K, Nishino, T. | | Deposit date: | 2011-03-31 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase
J.Am.Chem.Soc., 134, 2012
|
|
2EX2
 
 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli | | Descriptor: | GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-07 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EXA
 
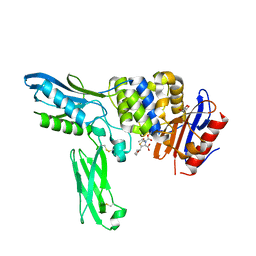 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with FAROM | | Descriptor: | (2R,5R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydro-1,3-thiazole-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-06-13 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EX8
 
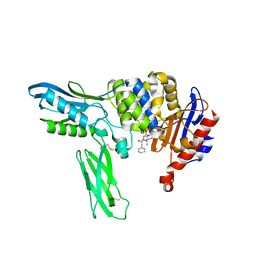 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with penicillin-G | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EX9
 
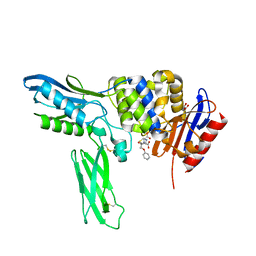 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with penicillin-V | | Descriptor: | (2R,4S)-5,5-dimethyl-2-{(1R)-2-oxo-1-[(phenoxyacetyl)amino]ethyl}-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-06-13 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EXB
 
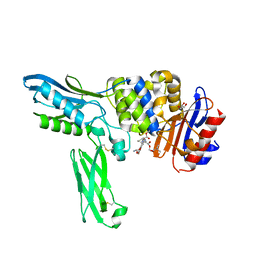 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with FLOMOX | | Descriptor: | 2,2-dimethylpropanoyloxymethyl (2R)-5-(aminocarbonyloxymethyl)-2-[(1R)-1-[[(Z)-2-(2-azanyl-1,3-thiazol-4-yl)pent-2-enoyl]amino]-2-oxidanylidene-ethyl]- 3,6-dihydro-2H-1,3-thiazine-4-carboxylate, GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
3A4Y
 
 | | Crystal Structure of H61A mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of H61A mutant TTHA0252 from Thermus thermophilus HB8
to be published
|
|
5UZI
 
 | | Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A - A6-DNAm1A16 structure | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*TP*TP*TP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*(M1A)P*AP*AP*AP*AP*AP*TP*CP*G)-3') | | Authors: | Sathyamoorthy, B, Shi, H, Xue, Y, Al-Hashimi, H.M. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A.
Nucleic Acids Res., 45, 2017
|
|
