8HDD
 
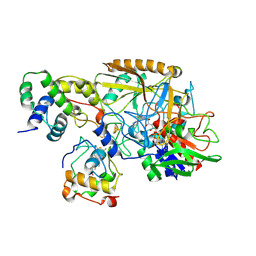 | | Complex structure of catalytic, small, and a partial electron transfer subunits from Burkholderia cepacia FAD glucose dehydrogenase | | 分子名称: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Glucose dehydrogenase, ... | | 著者 | Yoshida, H, Sode, K. | | 登録日 | 2022-11-04 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Microgravity environment grown crystal structure information based engineering of direct electron transfer type glucose dehydrogenase.
Commun Biol, 5, 2022
|
|
1IQC
 
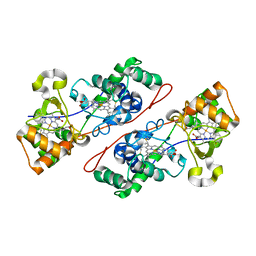 | |
3IEK
 
 | | Crystal Structure of native TTHA0252 from Thermus thermophilus HB8 | | 分子名称: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | 著者 | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R. | | 登録日 | 2009-07-22 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal Structure of native TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IEL
 
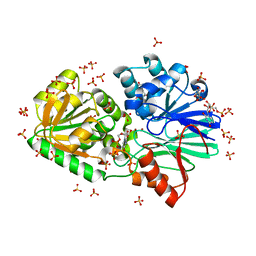 | | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with UMP | | 分子名称: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | 著者 | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R. | | 登録日 | 2009-07-23 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with UMP
To be Published
|
|
3IE1
 
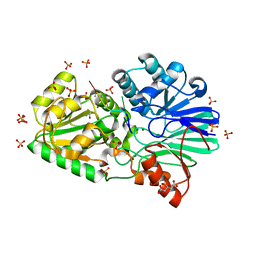 | | Crystal structure of H380A mutant TTHA0252 from Thermus thermophilus HB8 complexed with RNA | | 分子名称: | CITRATE ANION, RNA (5'-R(P*UP*UP*UP*U)-3'), Ribonuclease TTHA0252, ... | | 著者 | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-07-22 | | 公開日 | 2009-08-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal structure of H380A mutant TTHA0252 from Thermus thermophilus HB8 complexed with RNA
To be Published
|
|
3IE2
 
 | | Crystal Structure of H400V mutant TTHA0252 from Thermus thermophilus HB8 | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ribonuclease TTHA0252, SULFATE ION, ... | | 著者 | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-07-22 | | 公開日 | 2009-08-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of H400V mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
2RSD
 
 | | Solution structure of the plant homeodomain (PHD) of the E3 SUMO ligase Siz1 from rice | | 分子名称: | E3 SUMO-protein ligase SIZ1, ZINC ION | | 著者 | Shindo, H, Tsuchiya, W, Suzuki, R, Yamazaki, T. | | 登録日 | 2012-01-12 | | 公開日 | 2012-08-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | PHD finger of the SUMO ligase Siz/PIAS family in rice reveals specific binding for methylated histone H3 at lysine 4 and arginine 2
Febs Lett., 586, 2012
|
|
3IDZ
 
 | | Crystal Structure of S378Q mutant TTHA0252 from Thermus thermophilus HB8 | | 分子名称: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | 著者 | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-07-22 | | 公開日 | 2009-08-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of S378Q mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IE0
 
 | | Crystal Structure of S378Y mutant TTHA0252 from Thermus thermophilus HB8 | | 分子名称: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | 著者 | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-07-22 | | 公開日 | 2009-08-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Crystal Structure of S378Y mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IEM
 
 | | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with RNA analog | | 分子名称: | CITRATE ANION, RNA (5'-R(*(SSU)P*(SSU)P*(SSU)P*(SSU)P*(SSU)P*(SSU))-3'), Ribonuclease TTHA0252, ... | | 著者 | Ishikawa, H, Nakagawa, N, Kuramitus, S, Yokoyama, S, Masui, R. | | 登録日 | 2009-07-23 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with RNA analog
To be Published
|
|
7CJ4
 
 | |
7CJ9
 
 | | Crystal structure of N-terminal His-tagged D-allulose 3-epimerase from Methylomonas sp. with additional C-terminal residues | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-fructose, ... | | 著者 | Yoshida, H, Yoshihara, A, Kamitori, S. | | 登録日 | 2020-07-09 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
7CJ8
 
 | | Crystal structure of N-terminal His-tagged D-allulose 3-epimerase from Methylomonas sp. in complex with D-allulose | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-psicose, Epimerase, ... | | 著者 | Yoshida, H, Yoshihara, A, Kamitori, S. | | 登録日 | 2020-07-09 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
7CJ6
 
 | |
7CJ7
 
 | | Crystal structure of homo dimeric D-allulose 3-epimerase from Methylomonas sp. in complex with L-tagatose | | 分子名称: | (2R,3R,4R,5S)-2-(hydroxymethyl)oxane-2,3,4,5-tetrol, Epimerase, L-sorbose, ... | | 著者 | Yoshida, H, Yoshihara, A, Kamitori, S. | | 登録日 | 2020-07-09 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.695 Å) | | 主引用文献 | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
7CJ5
 
 | | Crystal structure of homo dimeric D-allulose 3-epimerase from Methylomonas sp. in complex with D-fructose | | 分子名称: | D-fructose, Epimerase, MAGNESIUM ION, ... | | 著者 | Yoshida, H, Yoshihara, A, Kamitori, S. | | 登録日 | 2020-07-09 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
7V3D
 
 | |
4JGC
 
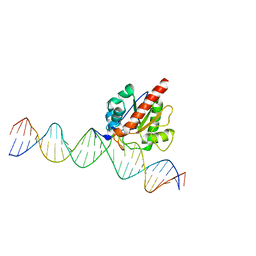 | | Human TDG N140A mutant IN A COMPLEX WITH 5-carboxylcytosine (5caC) | | 分子名称: | 4-amino-2-oxo-1,2-dihydropyrimidine-5-carboxylic acid, G/T mismatch-specific thymine DNA glycosylase, oligonucleotide, ... | | 著者 | Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2013-02-28 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.582 Å) | | 主引用文献 | Activity and crystal structure of human thymine DNA glycosylase mutant N140A with 5-carboxylcytosine DNA at low pH.
Dna Repair, 12, 2013
|
|
1NDH
 
 | |
1KXW
 
 | |
4LT5
 
 | | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA, MANGANESE (II) ION, ... | | 著者 | Hashimoto, H, Pais, J.E, Zhang, X, Saleh, L, Fu, Z.Q, Dai, N, Correa, I.R, Roberts, R.J, Zheng, Y, Cheng, X. | | 登録日 | 2013-07-23 | | 公開日 | 2013-12-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.893 Å) | | 主引用文献 | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA.
Nature, 506, 2014
|
|
6LEH
 
 | |
1KXX
 
 | |
1KXY
 
 | |
2EX6
 
 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with ampicillin | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4 | | 著者 | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2005-11-08 | | 公開日 | 2006-06-13 | | 最終更新日 | 2016-10-19 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
