1JFC
 
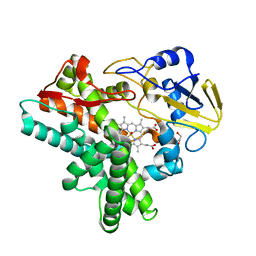 | | X-ray structure of nitric oxide reductase (cytochrome P450nor) in the ferrous CO state at atomic resolution | | Descriptor: | CARBON MONOXIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shimizu, H, Adachi, S, Park, S.Y, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-06-20 | | Release date: | 2001-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | X-ray structure of nitric oxide reductase (cytochrome P450nor) at atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1MGT
 
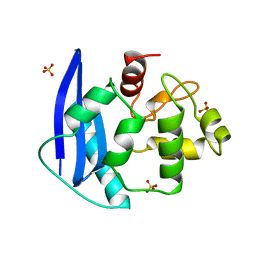 | | CRYSTAL STRUCTURE OF O6-METHYLGUANINE-DNA METHYLTRANSFERASE FROM HYPERTHERMOPHILIC ARCHAEON PYROCOCCUS KODAKARAENSIS STRAIN KOD1 | | Descriptor: | PROTEIN (O6-METHYLGUANINE-DNA METHYLTRANSFERASE), SULFATE ION | | Authors: | Hashimoto, H, Inoue, T, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 1999-01-12 | | Release date: | 2000-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hyperthermostable protein structure maintained by intra and inter-helix ion-pairs in archaeal O6-methylguanine-DNA methyltransferase.
J.Mol.Biol., 292, 1999
|
|
5XAX
 
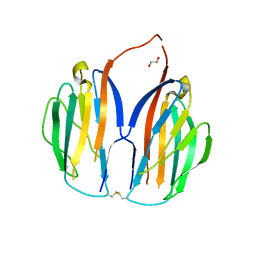 | |
5XAW
 
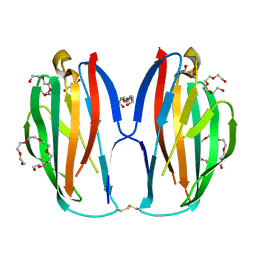 | | Parallel homodimer structures of voltage-gated sodium channel beta4 for cell-cell adhesion | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, GLYCEROL, Sodium channel subunit beta-4, ... | | Authors: | Shimizu, H, Yokoyama, S. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Parallel homodimer structures of the extracellular domains of the voltage-gated sodium channel beta 4 subunit explain its role in cell-cell adhesion
J. Biol. Chem., 292, 2017
|
|
6CPJ
 
 | | Solution structure of SH3 domain from Shank2 | | Descriptor: | SH3 and multiple ankyrin repeat domains protein 2 | | Authors: | Ishida, H, Vogel, H.J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domains from Shank scaffold proteins and their interactions with Cav1.3 calcium channels.
FEBS Lett., 592, 2018
|
|
6CPK
 
 | | Solution structure of SH3 domain from Shank3 | | Descriptor: | SH3 and multiple ankyrin repeat domains protein 3 | | Authors: | Ishida, H, Vogel, H.J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domains from Shank scaffold proteins and their interactions with Cav1.3 calcium channels.
FEBS Lett., 592, 2018
|
|
6CPI
 
 | | Solution structure of SH3 domain from Shank1 | | Descriptor: | SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Ishida, H, Vogel, H.J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the SH3 domains from Shank scaffold proteins and their interactions with Cav1.3 calcium channels.
FEBS Lett., 592, 2018
|
|
5AYQ
 
 | |
7CJ9
 
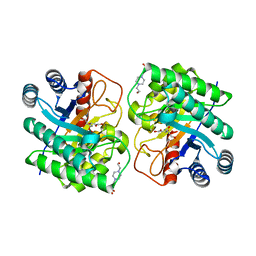 | | Crystal structure of N-terminal His-tagged D-allulose 3-epimerase from Methylomonas sp. with additional C-terminal residues | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-fructose, ... | | Authors: | Yoshida, H, Yoshihara, A, Kamitori, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
6DNZ
 
 | | Trypanosoma brucei PRMT1 enzyme-prozyme heterotetrameric complex with AdoHcy | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arginine N-methyltransferase, putative, ... | | Authors: | Hashimoto, H, Kafkova, L, Jordan, K, Read, L.K, Debler, E.W. | | Deposit date: | 2018-06-08 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | Structural Basis of Protein Arginine Methyltransferase Activation by a Catalytically Dead Homolog (Prozyme).
J.Mol.Biol., 432, 2020
|
|
7CJ8
 
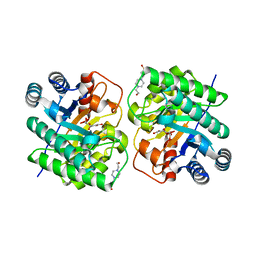 | | Crystal structure of N-terminal His-tagged D-allulose 3-epimerase from Methylomonas sp. in complex with D-allulose | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-psicose, Epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Kamitori, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
7CJ6
 
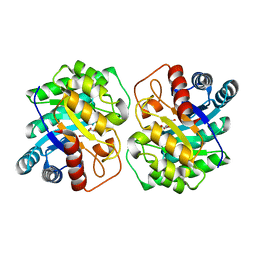 | |
7CJ7
 
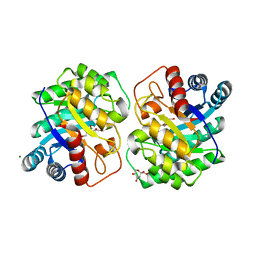 | | Crystal structure of homo dimeric D-allulose 3-epimerase from Methylomonas sp. in complex with L-tagatose | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)oxane-2,3,4,5-tetrol, Epimerase, L-sorbose, ... | | Authors: | Yoshida, H, Yoshihara, A, Kamitori, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
7CJ5
 
 | | Crystal structure of homo dimeric D-allulose 3-epimerase from Methylomonas sp. in complex with D-fructose | | Descriptor: | D-fructose, Epimerase, MAGNESIUM ION, ... | | Authors: | Yoshida, H, Yoshihara, A, Kamitori, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
1LKJ
 
 | | NMR Structure of Apo Calmodulin from Yeast Saccharomyces cerevisiae | | Descriptor: | Calmodulin | | Authors: | Ishida, H, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | Deposit date: | 2002-04-25 | | Release date: | 2003-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of apocalmodulin from Saccharomyces cerevisiae implies a mechanism for its unique Ca2+ binding property.
Biochemistry, 41, 2002
|
|
4YNT
 
 | | Crystal structure of Aspergillus flavus FAD glucose dehydrogenase | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Glucose oxidase, putative | | Authors: | Yoshida, H, Sakai, G, Kojima, K, Kamitori, S, Sode, K. | | Deposit date: | 2015-03-11 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural analysis of fungus-derived FAD glucose dehydrogenase
Sci Rep, 5, 2015
|
|
4YNU
 
 | | Crystal structure of Aspergillus flavus FADGDH in complex with D-glucono-1,5-lactone | | Descriptor: | D-glucono-1,5-lactone, FLAVIN-ADENINE DINUCLEOTIDE, Glucose oxidase, ... | | Authors: | Yoshida, H, Sakai, G, Kojima, K, Kamitori, S, Sode, K. | | Deposit date: | 2015-03-11 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural analysis of fungus-derived FAD glucose dehydrogenase
Sci Rep, 5, 2015
|
|
8HN3
 
 | | Soluble domain of cytochrome c-556 from Chlorobaculum tepidum | | Descriptor: | ACETATE ION, Cytochrome c-556, GLYCEROL, ... | | Authors: | Kishimoto, H, Azai, C, Yamamoto, T, Mutoh, R, Nakaniwa, T, Tanaka, H, Kurisu, G, Oh-oka, H. | | Deposit date: | 2022-12-07 | | Release date: | 2023-07-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Soluble domains of cytochrome c-556 and Rieske iron-sulfur protein from Chlorobaculum tepidum: Crystal structures and interaction analysis.
Curr Res Struct Biol, 5, 2023
|
|
3ROC
 
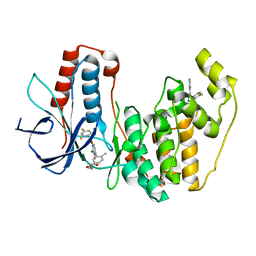 | | Crystal structure of human p38 alpha complexed with a pyrimidinone compound | | Descriptor: | 3-{5-chloro-4-[(2,4-difluorobenzyl)oxy]-6-oxopyrimidin-1(6H)-yl}-N-(2-hydroxyethyl)-4-methylbenzamide, 4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Shieh, H.-S, Xing, L. | | Deposit date: | 2011-04-25 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substituted N-aryl-6-pyrimidinones: A new class of potent, selective, and orally active p38 MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7CJ4
 
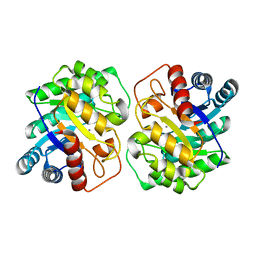 | |
1C8I
 
 | | BINDING MODE OF HYDROXYLAMINE TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HYDROXYAMINE, ... | | Authors: | Wariishi, H, Nonaka, D, Johjima, T, Nakamura, N, Naruta, Y, Kubo, K, Fukuyama, K. | | Deposit date: | 2000-05-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct binding of hydroxylamine to the heme iron of Arthromyces ramosus peroxidase. Substrate analogue that inhibits compound I formation in a competetive manner.
J.Biol.Chem., 275, 2000
|
|
8HN2
 
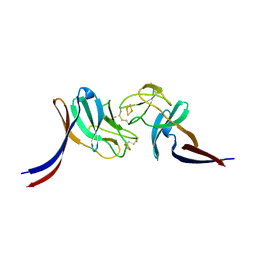 | | Selenomethionine-labelled soluble domain of Rieske iron-sulfur protein from chlorobaculum tepidum | | Descriptor: | Cytochrome b6-f complex iron-sulfur subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Kishimoto, H, Mutoh, R, Tanaka, H, Kurisu, G, Oh-oka, H. | | Deposit date: | 2022-12-07 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Soluble domains of cytochrome c-556 and Rieske iron-sulfur protein from Chlorobaculum tepidum: Crystal structures and interaction analysis.
Curr Res Struct Biol, 5, 2023
|
|
3HP5
 
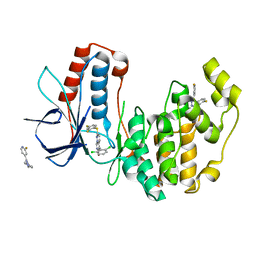 | | Crystal Structure of Human p38alpha complexed with a pyrimidopyridazinone compound | | Descriptor: | 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, 5-(2,6-dichlorophenyl)-2-[(2,4-difluorophenyl)sulfanyl]-6H-pyrimido[1,6-b]pyridazin-6-one, Mitogen-activated protein kinase 14 | | Authors: | Shieh, H.-S, Williams, J.M, Stegeman, R.A, Kurumbail, R.G. | | Deposit date: | 2009-06-03 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of N-substituted pyridinones as potent and selective inhibitors of p38 kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3HY7
 
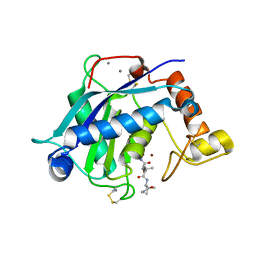 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with Marimastat | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors
J.Biol.Chem., 284, 2009
|
|
2ROA
 
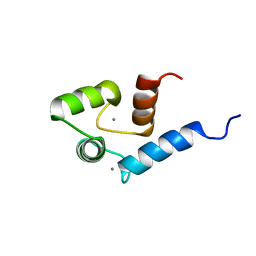 | | Solution structure of calcium bound soybean calmodulin isoform 4 N-terminal domain | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ishida, H, Huang, H, Yamniuk, A.P, Takaya, Y, Vogel, H.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties
J.Biol.Chem., 283, 2008
|
|
