6PZA
 
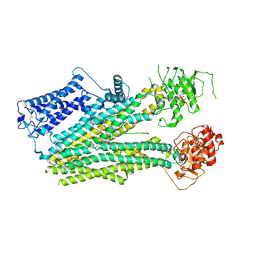 | | Cryo-EM structure of the pancreatic beta-cell SUR1 bound to ATP and glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Shyng, S.L, Yoshioka, C, Martin, G.M, Sung, M.W. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Mechanism of pharmacochaperoning in a mammalian K ATP channel revealed by cryo-EM.
Elife, 8, 2019
|
|
5KK2
 
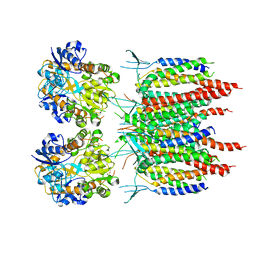 | | Architecture of fully occupied GluA2 AMPA receptor - TARP complex elucidated by single particle cryo-electron microscopy | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Zhao, Y, Chen, S, Yoshioka, C, Baconguis, I, Gouaux, E. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM.
Nature, 536, 2016
|
|
6PZ9
 
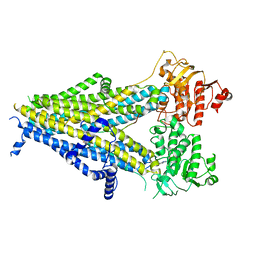 | | Cryo-EM structure of the pancreatic beta-cell SUR1 bound to ATP and repaglinide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ATP-sensitive inward rectifier potassium channel 11, ... | | Authors: | Shyng, S.L, Yoshioka, C, Martin, G.M, Sung, M.W. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Mechanism of pharmacochaperoning in a mammalian K ATP channel revealed by cryo-EM.
Elife, 8, 2019
|
|
6PZC
 
 | |
5XQU
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20I mutant (NFE6, AFP), P212121 form | | Descriptor: | Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XQP
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein (NFE6, AFP), P212121 form | | Descriptor: | Ice-structuring protein, SULFATE ION | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8IC1
 
 | | endo-alpha-D-arabinanase EndoMA1 D51N mutant from Microbacterium arabinogalactanolyticum in complex with arabinooligosaccharides | | Descriptor: | (3~{a}~{S},5~{R},6~{R},6~{a}~{S})-5-(hydroxymethyl)-2,2-dimethyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]dioxol-6-ol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Li, J, Nakashima, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
6NZ0
 
 | | Cryo-EM structure of AAV-2 in complex with AAVR PKD domains 1 and 2 | | Descriptor: | Capsid protein VP1, Dyslexia-associated protein KIAA0319-like protein, MAGNESIUM ION | | Authors: | Meyer, N.L, Xie, Q, Davulcu, O, Yoshioka, C, Chapman, M.S. | | Deposit date: | 2019-02-12 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of the gene therapy vector, adeno-associated virus with its cell receptor, AAVR.
Elife, 8, 2019
|
|
5XAA
 
 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E2-ALF-(TG) crystals of P21212 symmetry | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
5XA8
 
 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E1-ALF4-ADP-2CA2+ crystals | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
5XA9
 
 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E2-ALF-(TG) crystals of C2 symmetry | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
5XA7
 
 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E1-2CA2+ crystals | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, SODIUM ION, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
1IS4
 
 | | LACTOSE-LIGANDED CONGERIN II | | Descriptor: | CONGERIN II, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Conger Eel Galectin (Congerin II) at 1.45 A Resolution: Implication for the Accelerated Evolution of a New Ligand-Binding Site Following Gene Duplication
J.Mol.Biol., 321, 2002
|
|
1IS3
 
 | | LACTOSE AND MES-LIGANDED CONGERIN II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CONGERIN II, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a conger eel galectin (congerin II) at 1.45 A resolution: Implication for the accelerated evolution of a new ligand-binding site following gene duplication
J.MOL.BIOL., 321, 2002
|
|
1IS5
 
 | | Ligand free Congerin II | | Descriptor: | Congerin II | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a conger eel galectin (congerin II) at 1.45 A resolution: Implication for the accelerated evolution of a new ligand-binding site following gene duplication
J.MOL.BIOL., 321, 2002
|
|
1IS6
 
 | | MES-Liganded Congerin II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Congerin II | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a conger eel galectin (congerin II) at 1.45 A resolution: Implication for the accelerated evolution of a new ligand-binding site following gene duplication
J.MOL.BIOL., 321, 2002
|
|
1C1F
 
 | | LIGAND-FREE CONGERIN I | | Descriptor: | PROTEIN (CONGERIN I) | | Authors: | Shirai, T, Mitsuyama, C, Niwa, Y, Matsui, Y, Hotta, H, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structure of the conger eel galectin, congerin I, in lactose-liganded and ligand-free forms: emergence of a new structure class by accelerated evolution.
Structure Fold.Des., 7, 1999
|
|
7BYD
 
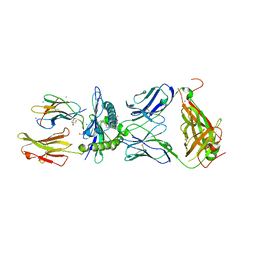 | | Crystal structure of SN45 TCR in complex with lipopeptide-bound Mamu-B*05104 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, B protein, ... | | Authors: | Morita, D, Sugita, M, Iwashita, C. | | Deposit date: | 2020-04-22 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.80003262 Å) | | Cite: | Crystal structure of the ternary complex of TCR, MHC class I and lipopeptides.
Int.Immunol., 32, 2020
|
|
1C1L
 
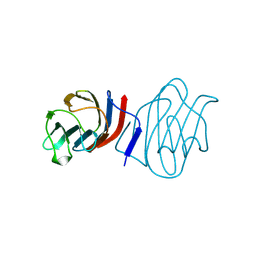 | | LACTOSE-LIGANDED CONGERIN I | | Descriptor: | PROTEIN (CONGERIN I), beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shirai, T, Mitsuyama, C, Niwa, Y, Matsui, Y, Hotta, H, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structure of the conger eel galectin, congerin I, in lactose-liganded and ligand-free forms: emergence of a new structure class by accelerated evolution.
Structure Fold.Des., 7, 1999
|
|
1J2E
 
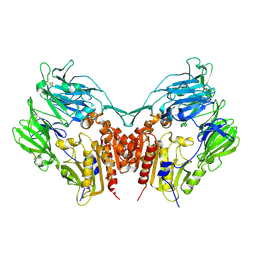 | | Crystal structure of Human Dipeptidyl peptidase IV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV | | Authors: | Hiramatsu, H, Kyono, K, Higashiyama, Y, Fukushima, C, Shima, H, Sugiyama, S, Inaka, K, Yamamoto, A, Shimizu, R. | | Deposit date: | 2002-12-30 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and function of human dipeptidyl peptidase IV, possessing a unique eight-bladed beta-propeller fold.
Biochem.Biophys.Res.Commun., 302, 2003
|
|
4YCN
 
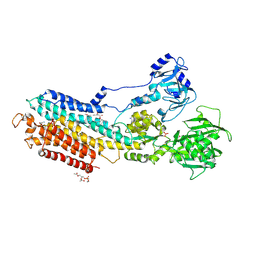 | | Crystal structure of the calcium pump with bound marine macrolide BLLB | | Descriptor: | (4S,5E,8S,9E,11S,13E,15E,18R)-4-hydroxy-8-methoxy-9,11-dimethyl-18-[(1Z,4E)-2-methylhexa-1,4-dien-1-yl]oxacyclooctadeca-5,9,13,15-tetraen-2-one, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Morita, M, Ogawa, H, Ohno, O, Yamori, T, Suenaga, K, Toyoshima, C. | | Deposit date: | 2015-02-20 | | Release date: | 2016-01-13 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Biselyngbyasides, cytotoxic marine macrolides, are novel and potent inhibitors of the Ca(2+) pumps with a unique mode of binding
Febs Lett., 589, 2015
|
|
4YCM
 
 | | Crystal structure of the calcium pump with bound marine macrolide BLS | | Descriptor: | (4S,5E,8S,9E,11S,13E,15E,18R)-8-methoxy-9,11-dimethyl-18-[(1Z,4E)-2-methylhexa-1,4-dien-1-yl]-2-oxooxacyclooctadeca-5,9,13,15-tetraen-4-yl 3-O-methyl-beta-D-glucopyranoside, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Morita, M, Ogawa, H, Ohno, O, Yamori, T, Suenaga, K, Toyoshima, C. | | Deposit date: | 2015-02-20 | | Release date: | 2016-01-13 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biselyngbyasides, cytotoxic marine macrolides, are novel and potent inhibitors of the Ca(2+) pumps with a unique mode of binding
Febs Lett., 589, 2015
|
|
5AW0
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 55 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AVV
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 8.5 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AW5
 
 | | Kinetics by X-ray crystallography: Rb+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 2.2 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
