1C1F
 
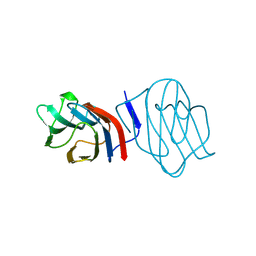 | | LIGAND-FREE CONGERIN I | | Descriptor: | PROTEIN (CONGERIN I) | | Authors: | Shirai, T, Mitsuyama, C, Niwa, Y, Matsui, Y, Hotta, H, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structure of the conger eel galectin, congerin I, in lactose-liganded and ligand-free forms: emergence of a new structure class by accelerated evolution.
Structure Fold.Des., 7, 1999
|
|
1YH3
 
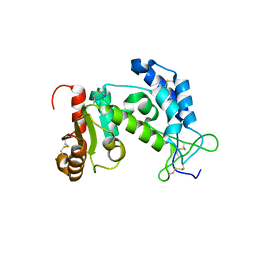 | | Crystal structure of human CD38 extracellular domain | | Descriptor: | ADP-ribosyl cyclase 1 | | Authors: | Liu, Q, Kriksunov, I.A, Graeff, R, Munshi, C, Lee, H.C, Hao, Q. | | Deposit date: | 2005-01-06 | | Release date: | 2005-09-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of human CD38 extracellular domain.
Structure, 13, 2005
|
|
1Z97
 
 | | Human Carbonic Anhydrase III: Structural and Kinetic Study of Catalysis and Proton Transfer. | | Descriptor: | Carbonic anhydrase III, ZINC ION | | Authors: | Duda, D.M, Tu, C, Fisher, S.Z, An, H, Yoshioka, C, Govindasamy, L, Laipis, P.J, Agbandje-McKenna, M, Silverman, D.N, McKenna, R. | | Deposit date: | 2005-03-31 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Carbonic Anhydrase III: Structural and Kinetic Study of Catalysis and Proton Transfer
Biochemistry, 44, 2005
|
|
1Z93
 
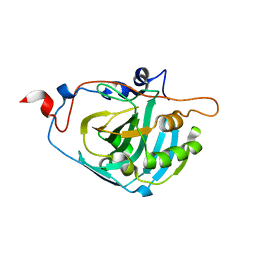 | | Human Carbonic Anhydrase III:Structural and Kinetic study of Catalysis and Proton Transfer. | | Descriptor: | Carbonic anhydrase III, ZINC ION | | Authors: | Duda, D.M, Tu, C, Fisher, S.Z, An, H, Yoshioka, C, Govindasamy, L, Laipis, P.J, Agbandje-McKenna, M, Silverman, D.N, McKenna, R. | | Deposit date: | 2005-03-31 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Carbonic Anhydrase III: Structural and Kinetic Study of Catalysis and Proton Transfer
Biochemistry, 44, 2005
|
|
1J2E
 
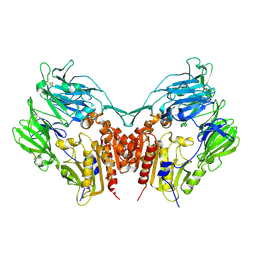 | | Crystal structure of Human Dipeptidyl peptidase IV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV | | Authors: | Hiramatsu, H, Kyono, K, Higashiyama, Y, Fukushima, C, Shima, H, Sugiyama, S, Inaka, K, Yamamoto, A, Shimizu, R. | | Deposit date: | 2002-12-30 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and function of human dipeptidyl peptidase IV, possessing a unique eight-bladed beta-propeller fold.
Biochem.Biophys.Res.Commun., 302, 2003
|
|
1PKF
 
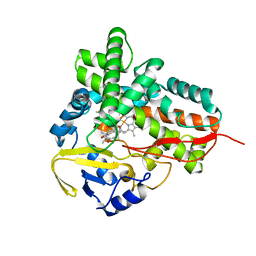 | | Crystal Structure of Epothilone D-bound Cytochrome P450epoK | | Descriptor: | EPOTHILONE D, PROTOPORPHYRIN IX CONTAINING FE, cytochrome p450EpoK | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-06-05 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Epothilone D-bound, Epothilone B-bound, and Substrate-free Forms of Cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
1Q5D
 
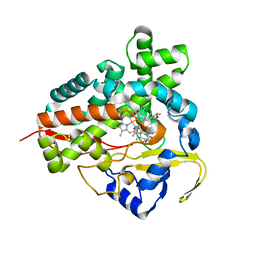 | | Epothilone B-bound Cytochrome P450epoK | | Descriptor: | 7,11-DIHYDROXY-8,8,10,12,16-PENTAMETHYL-3-[1-METHYL-2-(2-METHYL-THIAZOL-4-YL)VINYL]-4,17-DIOXABICYCLO[14.1.0]HEPTADECANE-5,9-DIONE, P450 epoxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
1C1L
 
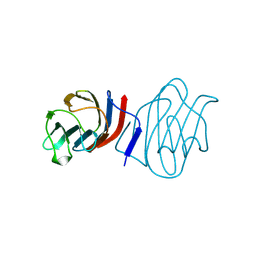 | | LACTOSE-LIGANDED CONGERIN I | | Descriptor: | PROTEIN (CONGERIN I), beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shirai, T, Mitsuyama, C, Niwa, Y, Matsui, Y, Hotta, H, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 1999-03-03 | | Release date: | 1999-10-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structure of the conger eel galectin, congerin I, in lactose-liganded and ligand-free forms: emergence of a new structure class by accelerated evolution.
Structure Fold.Des., 7, 1999
|
|
1Q5E
 
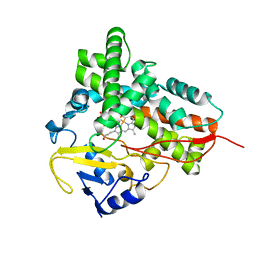 | | Substrate-free Cytochrome P450epoK | | Descriptor: | P450 epoxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
200D
 
 | | STABLE LOOP IN THE CRYSTAL STRUCTURE OF THE INTERCALATED FOUR-STRANDED CYTOSINE-RICH METAZOAN TELOMERE | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*CP*C)-3') | | Authors: | Kang, C, Berger, I, Lockshin, C, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1995-02-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Stable loop in the crystal structure of the intercalated four-stranded cytosine-rich metazoan telomere.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
7DCE
 
 | | Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Heavy chain of Fab fragment, Isoform 2 of Basigin, ... | | Authors: | Sakuragi, T, Kanai, R, Tsutsumi, A, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Kikkawa, M, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6BQN
 
 | | Cryo-EM structure of ENaC | | Descriptor: | 10D4 fab, 7B1 fab, EGFP-SCNN1G chimera, ... | | Authors: | Noreng, S, Bharadwaj, A, Posert, R, Yoshioka, C, Baconguis, I. | | Deposit date: | 2017-11-28 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the human epithelial sodium channel by cryo-electron microscopy.
Elife, 7, 2018
|
|
7JKC
 
 | | Sheep Connexin-46 at 1.9 angstroms resolution by CryoEM | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-28 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
3WGV
 
 | | Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state with oligomycin | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kanai, R, Ogawa, H, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a Na1-bound Na1,K1-ATPase preceding the E1P state
Nature, 502, 2013
|
|
3WGU
 
 | | Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state without oligomycin | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kanai, R, Ogawa, H, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a Na1-bound Na1,K1-ATPase preceding the E1P state
Nature, 502, 2013
|
|
7DAA
 
 | | Crystal structure of basigin complexed with anti-basigin Fab fragment | | Descriptor: | CADMIUM ION, Heavy chain of antibody Fab fragment, Isoform 2 of Basigin, ... | | Authors: | Sakuragi, T, Kanai, R, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-16 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JN0
 
 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 2 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-08-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JJP
 
 | | Sheep Connexin-50 at 1.9 angstroms resolution by CryoEM | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JN1
 
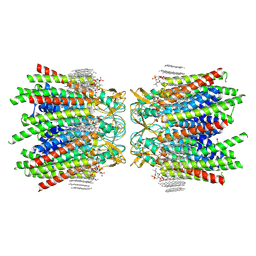 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-08-03 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JMD
 
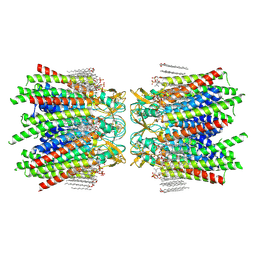 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 1 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JM9
 
 | | Sheep Connexin-50 at 2.5 angstroms reoslution, Lipid Class 2 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JLW
 
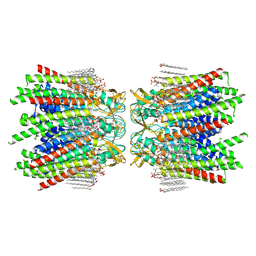 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 1 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
5ZMW
 
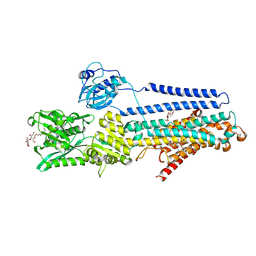 | | Crystal structure of the E309Q mutant of SR Ca2+-ATPase in E2(TG) | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Ogawa, H, Hirata, A, Tsueda, J, Toyoshima, C. | | Deposit date: | 2018-04-06 | | Release date: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of the E2 to E1 transition in Ca2+pump revealed by crystal structures of gating residue mutants.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZMV
 
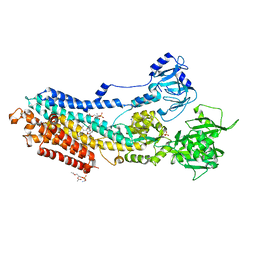 | | Crystal structure of the E309A mutant of SR Ca2+-ATPase in E2(TG) | | Descriptor: | OCTANOIC ACID [3S-[3ALPHA, 3ABETA, 4ALPHA, ... | | Authors: | Ogawa, H, Hirata, A, Tsueda, J, Toyoshima, C. | | Deposit date: | 2018-04-06 | | Release date: | 2018-12-12 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanism of the E2 to E1 transition in Ca2+pump revealed by crystal structures of gating residue mutants.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DW0
 
 | | Cryo-EM structure of the benzodiazepine-sensitive alpha1beta1gamma2S tri-heteromeric GABAA receptor in complex with GABA (Whole map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1,Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Phulera, S, Zhu, H, Yu, J, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the benzodiazepine-sensitive alpha 1 beta 1 gamma 2S tri-heteromeric GABAAreceptor in complex with GABA.
Elife, 7, 2018
|
|
