2YWE
 
 | | Crystal structure of LepA from Aquifex aeolicus | | Descriptor: | GTP-binding protein lepA | | Authors: | Kawazoe, M, Takemoto, C, Kaminishi, T, Nishino, A, Nakayama-Ushikoshi, R, Hanawa-Suetsugu, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of GTP-binding protein LepA from Aquifex aeolicus
To be Published
|
|
2YY1
 
 | | Crystal structure of N-terminal domain of human galectin-9 containing L-acetyllactosamine | | Descriptor: | Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of N-terminal domain of human galectin-9 containing L-acetyllactosamine
To be Published
|
|
2Z0W
 
 | | Crystal structure of the 2nd CAP-Gly domain in human Restin-like protein 2 reveals a swapped-dimer | | Descriptor: | CAP-Gly domain-containing linker protein 4 | | Authors: | Saito, K, Yoshikawa, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the 2nd CAP-Gly domain in human Restin-like protein 2 reveals a swapped-dimer
To be Published
|
|
8HHV
 
 | | endo-alpha-D-arabinanase EndoMA1 from Microbacterium arabinogalactanolyticum | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Nakashima, C, Li, J, Arakawa, T, Yamada, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2022-11-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
2Z0B
 
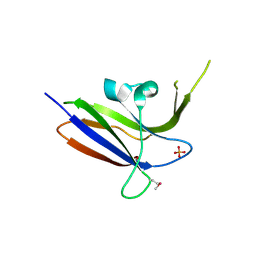 | | Crystal structure of CBM20 domain of human putative glycerophosphodiester phosphodiesterase 5 (KIAA1434) | | Descriptor: | PHOSPHATE ION, Putative glycerophosphodiester phosphodiesterase 5 | | Authors: | Saijo, S, Nishino, A, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of CBM20 domain of human putative glycerophosphodiester phosphodiesterase 5 (KIAA1434)
To be Published
|
|
4BIB
 
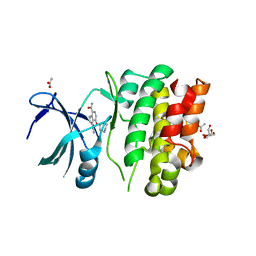 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 3-cyano-4-(piperidin-4-yloxy)-1H-indole-7-carboxamide, ACETATE ION, GLYCEROL, ... | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
8GYH
 
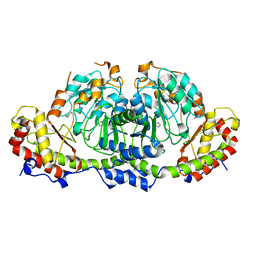 | | Crystal structure of Fic25 (apo form) from Streptomyces ficellus | | Descriptor: | DegT/DnrJ/EryC1/StrS family aminotransferase, GLYCEROL, IMIDAZOLE | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-09-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of Sugar Aminotransferase-like Enzymes to Synthesize Stereoisomers of Non-proteinogenic Amino Acids in Natural Product Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
8HPJ
 
 | |
8GYI
 
 | | Crystal structure of Fic25 (holo form) from Streptomyces ficellus | | Descriptor: | DegT/DnrJ/EryC1/StrS family aminotransferase, GLYCEROL, IMIDAZOLE, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-09-22 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanisms of Sugar Aminotransferase-like Enzymes to Synthesize Stereoisomers of Non-proteinogenic Amino Acids in Natural Product Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
8GYJ
 
 | | Crystal structure of Fic25 complexed with PLP-(5S,6S)-N2-acetyl-DADH adduct from Streptomyces ficellus | | Descriptor: | (2~{S},5~{S},6~{S})-2-acetamido-6-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-5,7-bis(oxidanyl)heptanoic acid, DegT/DnrJ/EryC1/StrS family aminotransferase, GLYCEROL, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-09-22 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Mechanisms of Sugar Aminotransferase-like Enzymes to Synthesize Stereoisomers of Non-proteinogenic Amino Acids in Natural Product Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
4BHN
 
 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 4-tert-butyl-N-[6-(1H-pyrazol-4-yl)imidazo[1,2-a]pyridin-2-yl]benzamide, GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5 | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
2YY9
 
 | | Crystal structure of BTB domain from mouse HKR3 | | Descriptor: | Zinc finger and BTB domain-containing protein 48 | | Authors: | Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of BTB domain from mouse HKR3
To be Published
|
|
4BF2
 
 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | ACETATE ION, GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, ... | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-03-13 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BID
 
 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 5-[(1R)-2-amino-1-phenylethoxy]-2-(furan-3-yl)-3-{1H-pyrrolo[2,3-b]pyridin-3-yl}pyridine, ACETATE ION, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5 | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
2Z15
 
 | | Crystal structure of human Tob1 protein | | Descriptor: | Protein Tob1 | | Authors: | Saito, K, Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human Tob1 protein
To be Published
|
|
4BIC
 
 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, N-(2-aminoethyl)-5-{1H-pyrrolo[2,3-b]pyridin-3-yl}thiophene-2-sulfonamide | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
8H20
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with Glycine | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H21
 
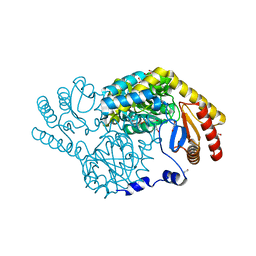 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-alanine | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H1Q
 
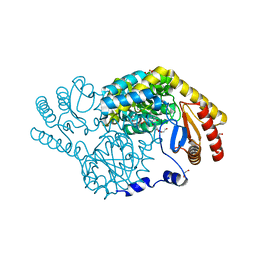 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-serine | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-03 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H29
 
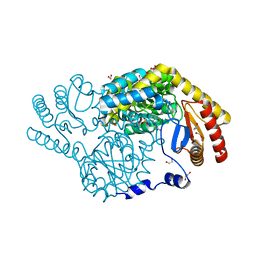 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-threonine | | Descriptor: | 1,2-ETHANEDIOL, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-threonine, Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-05 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H1Y
 
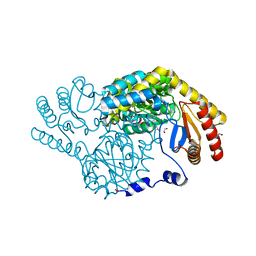 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-homoserine | | Descriptor: | (2~{S})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-oxidanyl-butanoic acid, 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
2Z17
 
 | | Crystal structure of PDZ domain from human Pleckstrin homology, Sec7 | | Descriptor: | Pleckstrin homology Sec7 and coiled-coil domains-binding protein | | Authors: | Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-08 | | Release date: | 2008-05-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of PDZ domain from human Pleckstrin homology, Sec7
To be Published
|
|
8HPK
 
 | | Crystal structure of the bacterial oxalate transporter OxlT in an oxalate-bound occluded form | | Descriptor: | Fab fragment Heavy chein, Fab fragment Light chain, OXALATE ION, ... | | Authors: | Shimamura, T, Hirai, T, Yamashita, A. | | Deposit date: | 2022-12-12 | | Release date: | 2023-02-15 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanism of oxalate transporter OxlT in an oxalate-degrading bacterium in the gut microbiota.
Nat Commun, 14, 2023
|
|
2YWK
 
 | | Crystal structure of RRM-domain derived from human putative RNA-binding protein 11 | | Descriptor: | Putative RNA-binding protein 11 | | Authors: | Kawazoe, M, Takemoto, C, Kaminishi, T, Uchikubo-Kamo, T, Nishino, A, Morita, S, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of RRM-domain derived from human putative RNA-binding protein 11
To be Published
|
|
2Z13
 
 | | Crystal structure of the N-terminal DUF1126 in human EF-hand domain | | Descriptor: | EF-hand domain-containing family member C2 | | Authors: | Saito, K, Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the N-terminal DUF1126 in human EF-hand domain
To be Published
|
|
