8CIK
 
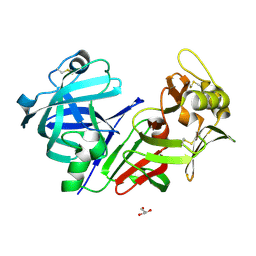 | | Altai wapiti (Cervus elaphus sibiricus) chymosin at 2.2 A resolution | | 分子名称: | Chymosin, GLYCEROL | | 著者 | Diusenova, S.E, Shevtsov, M.B, Borshchevskiy, V.I, Belenkaya, S.V, Kolybalov, D.S, Arkhipov, S.G, Volosnikova, E.A, Elchaninov, V.V, Shcherbakov, D.N. | | 登録日 | 2023-02-09 | | 公開日 | 2023-03-08 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Altai wapiti (Cervus elaphus sibiricus) chymosin at 2.2 A resolution
To Be Published
|
|
6J1K
 
 | | Crystal structure of ribose-5-phosphate isomerase A from Ochrobactrum sp. CSL1 | | 分子名称: | DI(HYDROXYETHYL)ETHER, Ribose-5-phosphate isomerase A | | 著者 | Xu, X.Q, Mo, X.B, Ju, X, Shen, M, Li, L.Z, Yuan, A.Y. | | 登録日 | 2018-12-28 | | 公開日 | 2019-04-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.824 Å) | | 主引用文献 | Crystal structure of ribose-5-phosphate isomerase A from Ochrobactrum sp. CSL1
To be published
|
|
6ODJ
 
 | | PolyAla Model of the PRC from the Type 4 Secretion System of H. pylori | | 分子名称: | PolyAla Model of PRC from H.pylori | | 著者 | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | 登録日 | 2019-03-26 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6OEG
 
 | | Structure of CagX from a cryo-EM reconstruction of a T4SS | | 分子名称: | Type IV secretion system apparatus protein CagX | | 著者 | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | 登録日 | 2019-03-27 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6OEH
 
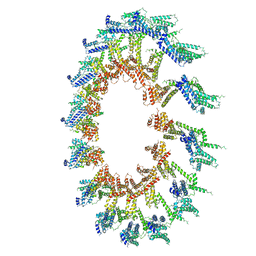 | | PolyAla Model of OMCC I-Layer | | 分子名称: | PolyAla Model of OMCC I-Layer | | 著者 | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | 登録日 | 2019-03-27 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6OEF
 
 | | PolyAla Model of the O-layer from the Type 4 Secretion System of H. pylori | | 分子名称: | PolyAla Model of OMCC O-Layer | | 著者 | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | 登録日 | 2019-03-27 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6ODI
 
 | | Structure of CagY from a cryo-EM reconstruction of a T4SS | | 分子名称: | Type IV secretion system apparatus protein CagY | | 著者 | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | 登録日 | 2019-03-26 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
5SVM
 
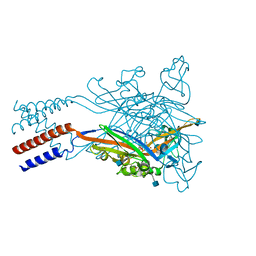 | | Crystal structure of the ATP-gated human P2X3 ion channel bound to agonist 2-methylthio-ATP in the desensitized state | | 分子名称: | 1,2-ETHANEDIOL, 2-(methylsulfanyl)adenosine 5'-(tetrahydrogen triphosphate), 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | 登録日 | 2016-08-06 | | 公開日 | 2016-09-28 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.093 Å) | | 主引用文献 | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5U4I
 
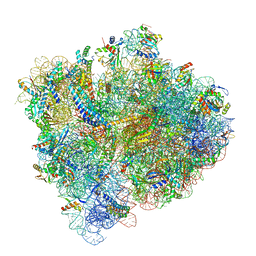 | | Structural Basis of Co-translational Quality Control by ArfA and RF2 Bound to Ribosome | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Zeng, F, Chen, Y, Remis, J, Shekhar, M, Phillips, J.C, Tajkhorshid, E, Jin, H. | | 登録日 | 2016-12-04 | | 公開日 | 2017-01-11 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome.
Nature, 541, 2017
|
|
5SVP
 
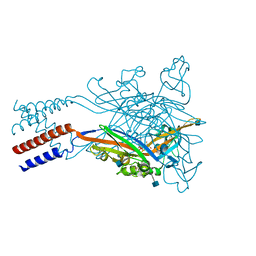 | | Anomalous sulfur signal reveals the position of agonist 2-methylthio-ATP bound to the ATP-gated human P2X3 ion channel in the desensitized state | | 分子名称: | 1,2-ETHANEDIOL, 2-(methylsulfanyl)adenosine 5'-(tetrahydrogen triphosphate), 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | 登録日 | 2016-08-07 | | 公開日 | 2016-09-28 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.298 Å) | | 主引用文献 | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5SVK
 
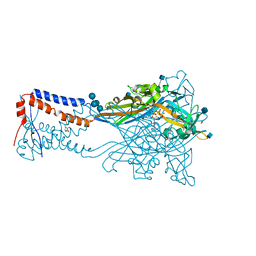 | | Crystal structure of the ATP-gated human P2X3 ion channel in the ATP-bound, open state | | 分子名称: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | 登録日 | 2016-08-06 | | 公開日 | 2016-09-28 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.773 Å) | | 主引用文献 | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5SVL
 
 | | Crystal structure of the ATP-gated human P2X3 ion channel in the ATP-bound, closed (desensitized) state | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | 登録日 | 2016-08-06 | | 公開日 | 2016-10-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
7K9P
 
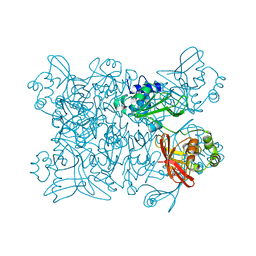 | | Room temperature structure of NSP15 Endoribonuclease from SARS CoV-2 solved using SFX. | | 分子名称: | CITRIC ACID, Uridylate-specific endoribonuclease | | 著者 | Botha, S, Jernigan, R, Chen, J, Coleman, M.A, Frank, M, Grant, T.D, Hansen, D.T, Ketawala, G, Logeswaran, D, Martin-Garcia, J, Nagaratnam, N, Raj, A.L.L.X, Shelby, M, Yang, J.-H, Yung, M.C, Fromme, P. | | 登録日 | 2020-09-29 | | 公開日 | 2020-10-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Room-temperature structural studies of SARS-CoV-2 protein NendoU with an X-ray free-electron laser.
Structure, 2022
|
|
6O2N
 
 | |
6O2M
 
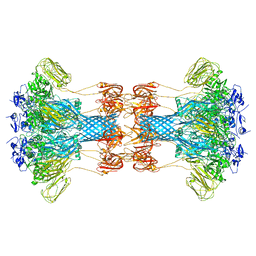 | |
6OKS
 
 | |
6O2O
 
 | |
6OKT
 
 | |
7VW7
 
 | | Crystal structure of the 2 ADP-AlF4-bound V1 complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Suzuki, K, Shekhar, M, Gupta, C, Singharoy, A, Murata, T. | | 登録日 | 2021-11-09 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.818 Å) | | 主引用文献 | Revealing a Hidden Intermediate of Rotatory Catalysis with X-ray Crystallography and Molecular Simulations.
Acs Cent.Sci., 8, 2022
|
|
6OKU
 
 | |
5FDL
 
 | | Crystal Structure of K103N/Y181C Mutant HIV-1 Reverse Transcriptase (RT) in Complex with IDX899 | | 分子名称: | P51 Reverse transcriptase, P66 Reverse transcriptase, methyl (R)-(2-carbamoyl-5-chloro-1H-indol-3-yl)[3-(2-cyanoethyl)-5-methylphenyl]phosphinate | | 著者 | Dousson, C.B, Alexandre, F.-R, Convard, T, Fisher, M, Lamers, M.B.A.C, Leonard, P.M. | | 登録日 | 2015-12-16 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Discovery of the Aryl-phospho-indole IDX899, a Highly Potent Anti-HIV Non-nucleoside Reverse Transcriptase Inhibitor.
J.Med.Chem., 59, 2016
|
|
5SVJ
 
 | | Crystal structure of the ATP-gated human P2X3 ion channel in the closed, apo state | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | 著者 | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | 登録日 | 2016-08-06 | | 公開日 | 2016-09-28 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.984 Å) | | 主引用文献 | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
6CXB
 
 | |
6CU2
 
 | |
5SVQ
 
 | | Crystal structure of the ATP-gated human P2X3 ion channel bound to competitive antagonist TNP-ATP | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, P2X purinoceptor 3, ... | | 著者 | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | 登録日 | 2016-08-07 | | 公開日 | 2016-10-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
