8C4I
 
 | |
8BC3
 
 | | Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex | | Descriptor: | (2~{R},3~{S},4~{S})-2,3,4,6-tetrakis(oxidanyl)hexane-1-sulfonic acid, BmSF-TAL | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8BC2
 
 | | Ligand-Free Structure of the decameric sulfofructose transaldolase BmSF-TAL | | Descriptor: | Transaldolase | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8BC4
 
 | | Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex in symmetry group C1 | | Descriptor: | (2~{R},3~{S},4~{S})-2,3,4,6-tetrakis(oxidanyl)hexane-1-sulfonic acid, Transaldolase | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
7NBZ
 
 | |
7OFY
 
 | | Crystal structure of SQ binding protein from Agrobacterium tumefaciens in complex with sulfoquinovosyl glycerol (SQGro) | | Descriptor: | 1,2-ETHANEDIOL, Sulfoquinovosyl binding protein, [(2S,3S,4S,5R,6S)-6-[(2R)-2,3-bis(oxidanyl)propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Jarva, M.A, Sharma, M, Goddard-Borger, E.D, Davies, G.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6ZEA
 
 | | Strictosidine Synthase from Catharanthus roseus in complex with racemic 1-methyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{R})-1-methyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, (1~{S})-1-methyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, 1,2-ETHANEDIOL, ... | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Strictosidine Synthase from Catharanthus roseus in complex with racemic 1-methyl-2,3,4,9-tetrahydro-1H-beta-carboline
To Be Published
|
|
6GMF
 
 | | Structure of Cytochrome P450 CYP109Q5 from Chondromyces apiculatus | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase | | Authors: | Grogan, G, Dubiel, P, Sharma, M, Klenk, J, Hauer, B. | | Deposit date: | 2018-05-25 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Characterization and structure-guided engineering of the novel versatile terpene monooxygenase CYP109Q5 from Chondromyces apiculatus DSM436.
Microb Biotechnol, 12, 2019
|
|
6G71
 
 | | Structure of CYP1232A24 from Arthrobacter sp. | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450, FE (III) ION, ... | | Authors: | Dubiel, P, Sharma, M, Klenk, J, Hauer, B, Grogan, G. | | Deposit date: | 2018-04-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and characterization of cytochrome P450 1232A24 and 1232F1 from Arthrobacter sp. and their role in the metabolic pathway of papaverine.
J.Biochem., 166, 2019
|
|
4UN2
 
 | | Crystal structure of the UBA domain of Dsk2 in complex with Ubiquitin | | Descriptor: | UBIQUITIN, UBIQUITIN DOMAIN-CONTAINING PROTEIN DSK2 | | Authors: | Michielssens, S, Peters, J.H, Ban, D, Pratihar, S, Seeliger, D, Sharma, M, Giller, K, Sabo, T.M, Becker, S, Lee, D, Griesinger, C, de Groot, B.L. | | Deposit date: | 2014-05-23 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A Designed Conformational Shift to Control Protein Binding Specificity.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
7BC0
 
 | |
7BBY
 
 | |
7BBZ
 
 | |
7BC1
 
 | |
7AO7
 
 | | Structure of CYP153A from Polaromonas sp. in complex with octan-1-ol | | Descriptor: | Cytochrome P450, OCTAN-1-OL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffmann, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
7ANT
 
 | | Structure of CYP153A from Polaromonas sp. | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450, HEME C | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffman, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-12 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
5FVB
 
 | | CRYSTAL STRUCTURE OF PHORMIDIUM C-PHYCOERYTHRIN AT PH 5.0 | | Descriptor: | C-PHYCOERYTHRIN ALPHA SUBUNIT, C-PHYCOERYTHRIN BETA SUBUNIT, GLYCEROL, ... | | Authors: | Kumar, V, Sonani, R.R, Sharma, M, Gupta, G.D, Madamwar, D. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure Analysis of C-Phycoerythrin from Marine Cyanobacterium Phormidium Sp. A09Dm.
Photosynth.Res., 129, 2016
|
|
8QC8
 
 | |
8QC2
 
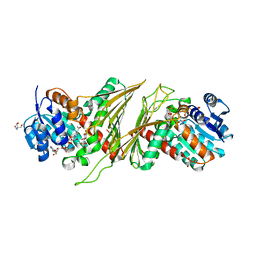 | | Crystal structure of NAD-dependent glycoside hydrolase from Flavobacterium sp. (strain K172) in complex with co-factor NAD+ and sulfoquinovose (SQ) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Pickles, I.B, Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Widespread Family of NAD + -Dependent Sulfoquinovosidases at the Gateway to Sulfoquinovose Catabolism.
J.Am.Chem.Soc., 145, 2023
|
|
6K5P
 
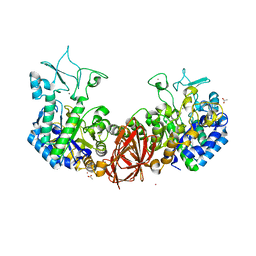 | | Structure of mosquito-larvicidal Binary toxin receptor, Cqm1 | | Descriptor: | ACETATE ION, Binary toxin receptor protein, CADMIUM ION, ... | | Authors: | Kumar, V, Sharma, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Crystal structure of BinAB toxin receptor (Cqm1) protein and molecular dynamics simulations reveal the role of unique Ca(II) ion.
Int.J.Biol.Macromol., 140, 2019
|
|
6S5J
 
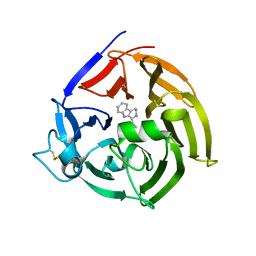 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with (S)-1-Ethyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{S})-1-ethyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Strictosidine synthase | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
6S5Q
 
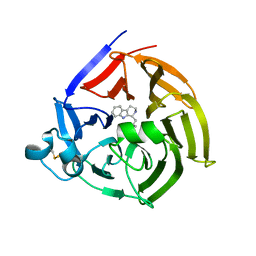 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with (S)-1-isobutyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{S})-1-(2-methylpropyl)-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Strictosidine synthase | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
6S5U
 
 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with N-[2-(1H-Indol-3-yl)ethyl]-3-methyl-1-butanamine | | Descriptor: | Strictosidine synthase, ~{N}-[2-(1~{H}-indol-3-yl)ethyl]-3-methyl-butan-1-amine | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
6S5M
 
 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with (S)-1-n-propyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{S})-1-propyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Strictosidine synthase | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
8FR8
 
 | | Structure of Mycobacterium smegmatis Rsh bound to a 70S translation initiation complex | | Descriptor: | 16S rRNA (1511-MER), 23S rRNA (3119-MER), 30S ribosomal protein S10, ... | | Authors: | Majumdar, S, Sharma, M.R, Manjari, S.R, Banavali, N.K, Agrawal, R.K. | | Deposit date: | 2023-01-06 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Starvation sensing by mycobacterial RelA/SpoT homologue through constitutive surveillance of translation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
