4Z86
 
 | |
5ZK0
 
 | | Crystal structure of Peptidyl-tRNA hydrolase mutant -M71A from Vibrio cholerae | | 分子名称: | Peptidyl-tRNA hydrolase, SODIUM ION | | 著者 | Shahid, S, Kabra, A, Pal, R.K, Arora, A. | | 登録日 | 2018-03-22 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Role of methionine 71 in substrate recognition and structural integrity of bacterial peptidyl-tRNA hydrolase.
Biochim. Biophys. Acta, 1866, 2018
|
|
6X9X
 
 | |
4ZXP
 
 | | Crystal structure of Peptidyl- tRNA Hydrolase from Vibrio cholerae | | 分子名称: | CITRATE ANION, Peptidyl-tRNA hydrolase | | 著者 | Shahid, S, Pal, R.K, Kabra, A, Yadav, R, Kumar, A, Arora, A. | | 登録日 | 2015-05-20 | | 公開日 | 2016-06-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Unraveling the stereochemical and dynamic aspects of the catalytic site of bacterial peptidyl-tRNA hydrolase.
RNA, 23, 2017
|
|
5B6J
 
 | |
5IKE
 
 | |
5IMB
 
 | |
5IVP
 
 | |
5GVZ
 
 | |
5EKT
 
 | |
2LME
 
 | | Solid-state NMR structure of the membrane anchor domain of the trimeric autotransporter YadA | | 分子名称: | Adhesin yadA | | 著者 | Shahid, S.A, Bardiaux, B, Franks, W.T, Habeck, M, Linke, D, van Rossum, B. | | 登録日 | 2011-11-30 | | 公開日 | 2012-11-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals.
Nat.Methods, 9, 2012
|
|
8TGV
 
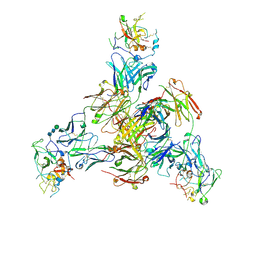 | | CryoEM structure of Fab HC84.26-HCV E2 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HC84.26 Heavy chain, ... | | 著者 | Shahid, S, Liqun, J, LIu, Y, Hasan, S.S, Mariuzza, R.A. | | 登録日 | 2023-07-13 | | 公開日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | CryoEM structure of Fab HC84.26-HCV E2 complex
To Be Published
|
|
8TGZ
 
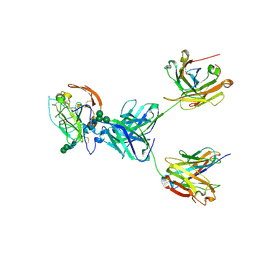 | | CryoEM structure of neutralizing antibody HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HC84.26 Heavy chain, ... | | 著者 | Shahid, S, Liqun, J, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | 登録日 | 2023-07-13 | | 公開日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | CryoEM structure of neutralizing antibody HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2
To Be Published
|
|
8THZ
 
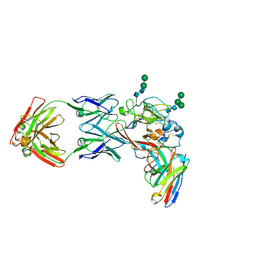 | | CryoEM structure of neutralizing antibodies CBH-7 and HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CBH-7 Heavy chain, ... | | 著者 | Shahid, S, Jiang, L, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | 登録日 | 2023-07-18 | | 公開日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | CryoEM structure of neutralizing antibodies CBH-7 and HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2
To Be Published
|
|
8TFE
 
 | |
4Z0X
 
 | |
2MJL
 
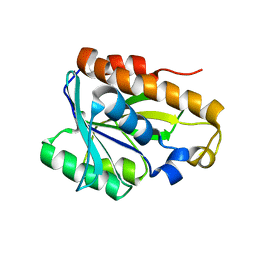 | | Solution structure of peptidyl-tRNA hyrolase from Vibrio cholerae | | 分子名称: | Peptidyl-tRNA hydrolase | | 著者 | Kabra, A, Shahid, S, Yadav, R, Pulavarti, S, Shukla, V.K, Arora, A. | | 登録日 | 2014-01-11 | | 公開日 | 2015-01-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of peptidyl-tRNA hydrolase from Vibrio cholerae
To be Published
|
|
8SO3
 
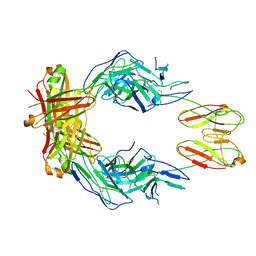 | | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte activation gene 3 protein, favezelimab Fab heavy chain, ... | | 著者 | Mishra, A.K, Shahid, S, Karade, S.S, Mariuzza, R.A. | | 登録日 | 2023-04-28 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.61 Å) | | 主引用文献 | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
8SR0
 
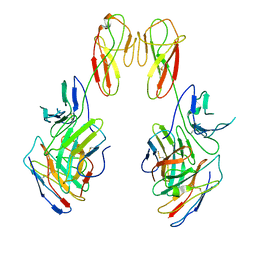 | | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 local refined | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte activation gene 3 protein, favezelimab Fab heavy chain, ... | | 著者 | Mishra, A.K, Shahid, S, Karade, S.S, Mariuzza, R.A. | | 登録日 | 2023-05-05 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
4C46
 
 | | ANDREI-N-LVPAS fused to GCN4 adaptors | | 分子名称: | BROMIDE ION, GENERAL CONTROL PROTEIN GCN4 | | 著者 | Albrecht, R, Alva, V, Ammelburg, M, Baer, K, Basina, E, Boichenko, I, Bonhoeffer, F, Braun, V, Chaubey, M, Chauhan, N, Chellamuthu, V.R, Coles, M, Deiss, S, Ewers, C.P, Forouzan, D, Fuchs, A, Groemping, Y, Hartmann, M.D, Hernandez Alvarez, B, Jeganantham, A, Kalev, I, Koenninger, U, Koiwai, K, Kopec, K.O, Korycinski, M, Laudenbach, B, Lehmann, K, Leo, J.C, Linke, D, Marialke, J, Martin, J, Mechelke, M, Michalik, M, Noll, A, Patzer, S.I, Scharfenberg, F, Schueckel, M, Shahid, S.A, Sulz, E, Ursinus, A, Wuertenberger, S, Zhu, H. | | 登録日 | 2013-08-30 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Your Personalized Protein Structure: Andrei N. Lupas Fused to GCN4 Adaptors.
J.Struct.Biol., 186, 2014
|
|
