7O5E
 
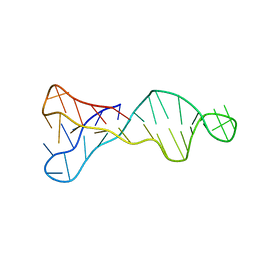 | |
8BQY
 
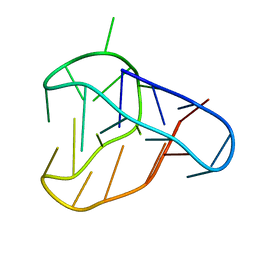 | | An i-motif domain able to undergo pH-dependent conformational transitions (acidic structure) | | 分子名称: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*(DNR)P*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*(DNR)P*CP*GP*T)-3') | | 著者 | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | 登録日 | 2022-11-22 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
8BV6
 
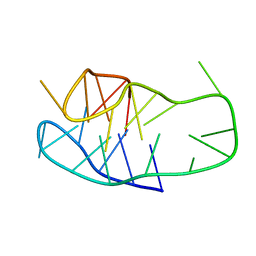 | | An i-motif domain able to undergo pH-dependent conformational transitions (neutral structure) | | 分子名称: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | 著者 | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | 登録日 | 2022-12-01 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
8OFC
 
 | | Structure of an i-motif domain with the cytosine analog 1,3-diaza-2-oxophenoxacione (tC) at neutral pH | | 分子名称: | DNA (5'-D(*CP*(YCO)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*(DNR)P*GP*T)-3') | | 著者 | Mir, B, Serrano-Chacon, I, Terrazas, M, Gandioso, A, Garavis, M, Orozco, M, Escaja, N, Gonzalez, C. | | 登録日 | 2023-03-15 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-04-24 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Site-specific incorporation of a fluorescent nucleobase analog enhances i-motif stability and allows monitoring of i-motif folding inside cells.
Nucleic Acids Res., 52, 2024
|
|
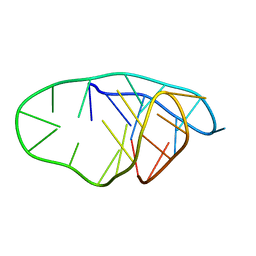
8PWR
 
 | | TINA-conjugated antiparallel DNA triplex | | 分子名称: | DNA (5'-D(*AP*GP*GP*AP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*TP*CP*CP*T)-3'), DNA (5'-D(*TP*GP*GP*TP*GP*(J32)P*GP*T)-3') | | 著者 | Garavis, M, Edwards, P.J.B, Serrano-Chacon, I, Doluca, O, Filichev, V.V, Gonzalez, C. | | 登録日 | 2023-07-21 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Understanding intercalative modulation of G-rich sequence folding: solution structure of a TINA-conjugated antiparallel DNA triplex.
Nucleic Acids Res., 52, 2024
|
|
