8FAL
 
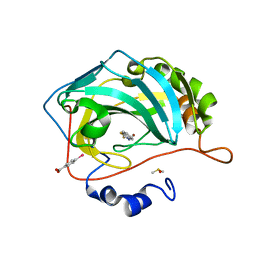 | | Masking thiol reactivity with thioamide-based MBPs- carbonic anhydrase II complexed with benzo[d]thiazole-2(3H)-thione | | Descriptor: | 1,3-benzothiazole-2(3H)-thione, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Kohlbrand, A.J, Seo, H. | | Deposit date: | 2022-11-28 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Masking thiol reactivity with thioamide, thiourea, and thiocarbamate-based MBPs.
Chem.Commun.(Camb.), 59, 2023
|
|
4OWR
 
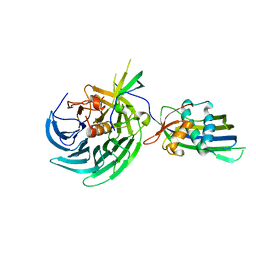 | | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair Rae1-Nup98 | | Descriptor: | Matrix protein, Nuclear pore complex protein Nup98-Nup96, mRNA export factor | | Authors: | Ren, Y, Quan, B, Seo, H.S, Blobel, G. | | Deposit date: | 2014-02-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 Nup98).
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8DAL
 
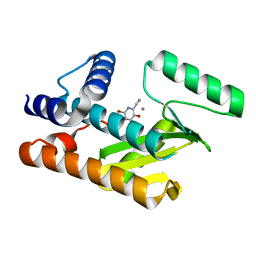 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 4-(benzyloxy)-6-bromo-3-hydroxypicolinonitrile | | Descriptor: | 6-bromo-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carbonitrile, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-13 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DDB
 
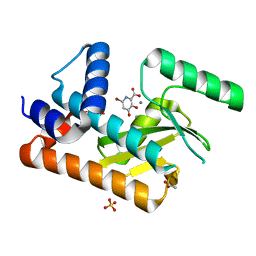 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 4-(benzyloxy)-6-bromo-3-hydroxypicolinic acid | | Descriptor: | 6-bromo-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DDE
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 4-(benzyloxy)-6-bromo-2-(1H-tetrazol-5-yl) yridine-3-ol | | Descriptor: | (2M)-6-bromo-3-hydroxy-2-(1H-tetrazol-5-yl)pyridin-4(1H)-one, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DHN
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 6-Bromo-3-hydroxy-2-(5-methyl-1,2,4-oxadiazol-3-yl)pyridin-4(1H)-one | | Descriptor: | (2P)-6-bromo-3-hydroxy-2-(5-methyl-1,2,4-oxadiazol-3-yl)pyridin-4(1H)-one, MANGANESE (II) ION, PA endonuclease | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-27 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8CTF
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid | | Descriptor: | 3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DJY
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 6-Bromo-2-(4,5-dihydro-1H-imidazol-2-yl)-3-hydroxypyridin-4(1H)-one | | Descriptor: | (2M)-6-bromo-2-(4,5-dihydro-1H-imidazol-2-yl)-3-hydroxypyridin-4(1H)-one, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-07-01 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DJV
 
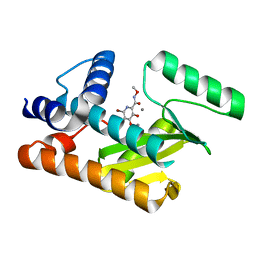 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 6-Bromo-3-hydroxy-N-methoxy-4-oxo-1,4-dihydropyridine-2-carboxamide | | Descriptor: | 6-bromo-3-hydroxy-N-methoxy-4-oxo-1,4-dihydropyridine-2-carboxamide, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-07-01 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
7V04
 
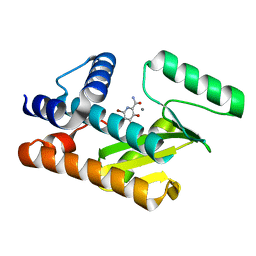 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 6-bromo-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxamide | | Descriptor: | 6-bromo-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxamide, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-05-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8HKB
 
 | |
8HKA
 
 | |
8HK9
 
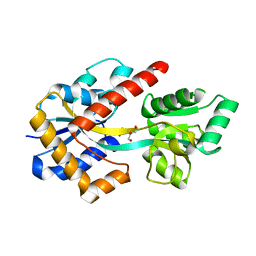 | |
7CWI
 
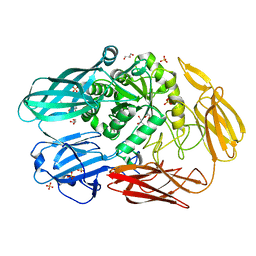 | | Crystal structure of beta-galactosidase II from Bacillus circulans | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Hong, H, Seo, H. | | Deposit date: | 2020-08-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High Galacto-Oligosaccharide Production and a Structural Model for Transgalactosylation of beta-Galactosidase II from Bacillus circulans .
J.Agric.Food Chem., 68, 2020
|
|
7CWD
 
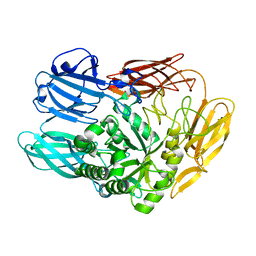 | |
7VG4
 
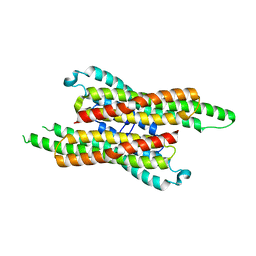 | | 10,5-methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1 strain | | Descriptor: | Methenyltetrahydrofolate cyclohydrolase | | Authors: | Kim, S, Lee, S, Kim, I.-K, Seo, H, Kim, K.-J. | | Deposit date: | 2021-09-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural insight into a molecular mechanism of methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1.
Int.J.Biol.Macromol., 202, 2022
|
|
7VG5
 
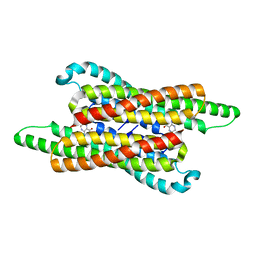 | | 10,5-methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1 with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, Methenyltetrahydrofolate cyclohydrolase | | Authors: | Kim, S, Lee, S, Kim, I.-K, Seo, H, Kim, K.-J. | | Deposit date: | 2021-09-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insight into a molecular mechanism of methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1.
Int.J.Biol.Macromol., 202, 2022
|
|
8H5M
 
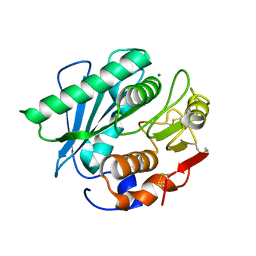 | |
8H5K
 
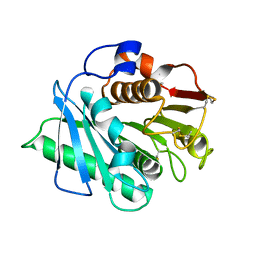 | |
8H5O
 
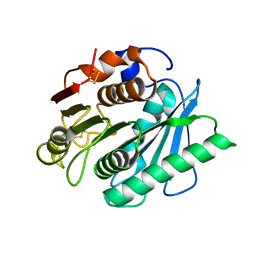 | |
8H5J
 
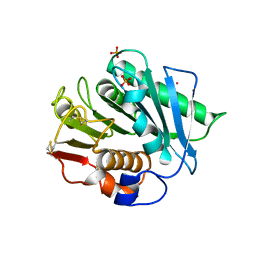 | |
8H5L
 
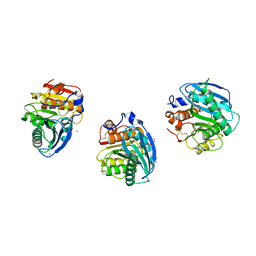 | |
6J57
 
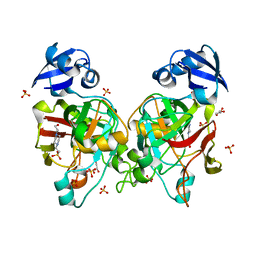 | | Crystal structure of fumarylpyruvate hydrolase from Corynebacterium glutamicum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Predicted 2-keto-4-pentenoate hydratase/2-oxohepta-3-ene-1,7-dioic acid hydratase, ... | | Authors: | Hong, H, Seo, H, Kim, K.-J, Park, W. | | Deposit date: | 2019-01-10 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Sequence, structure and function-based classification of the broadly conserved FAH superfamily reveals two distinct fumarylpyruvate hydrolase subfamilies.
Environ.Microbiol., 22, 2020
|
|
6J5Y
 
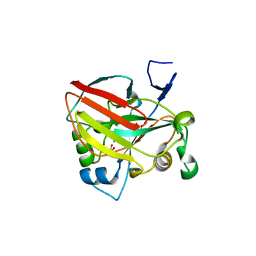 | | Crystal structure of fumarylpyruvate hydrolase from Pseudomonas aeruginosa in complex with Mn2+ and pyruvate | | Descriptor: | FAA hydrolase family protein, MANGANESE (II) ION, PYRUVIC ACID | | Authors: | Hong, H, Seo, H, Kim, K.-J, Park, W. | | Deposit date: | 2019-01-12 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Sequence, structure and function-based classification of the broadly conserved FAH superfamily reveals two distinct fumarylpyruvate hydrolase subfamilies.
Environ.Microbiol., 22, 2020
|
|
6J5X
 
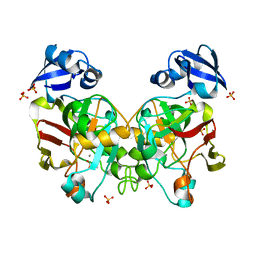 | | Crystal structure of fumarylpyruvate hydrolase from Corynebacterium glutamicum in complex with Mn2+ and pyruvate | | Descriptor: | MANGANESE (II) ION, PYRUVIC ACID, Predicted 2-keto-4-pentenoate hydratase/2-oxohepta-3-ene-1,7-dioic acid hydratase, ... | | Authors: | Hong, H, Seo, H, Kim, K.-J, Park, W. | | Deposit date: | 2019-01-12 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Sequence, structure and function-based classification of the broadly conserved FAH superfamily reveals two distinct fumarylpyruvate hydrolase subfamilies.
Environ.Microbiol., 22, 2020
|
|
