6MR4
 
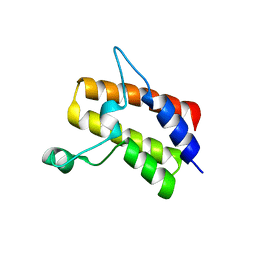 | | Crystal structure of the Sth1 bromodomain from S.cerevisiae | | Descriptor: | Nuclear protein STH1/NPS1 | | Authors: | Seo, H.S, Hashimoto, H, Krolak, A, Debler, E.W, Blus, B.J. | | Deposit date: | 2018-10-11 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Substrate Affinity and Specificity of the ScSth1p Bromodomain Are Fine-Tuned for Versatile Histone Recognition.
Structure, 27, 2019
|
|
6ISS
 
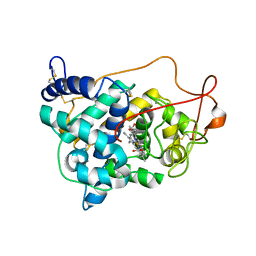 | | Lignin peroxidase H8 triple mutant S49C/A67C/H239 | | Descriptor: | CALCIUM ION, Ligninase H8, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Seo, H, Son, H, Kim, K.-J. | | Deposit date: | 2018-11-19 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Extra disulfide and ionic salt bridge improves the thermostability of lignin peroxidase H8 under acidic condition
Enzyme.Microb.Technol., 148, 2021
|
|
6IJK
 
 | |
6ITL
 
 | |
6ITK
 
 | |
3F7F
 
 | | Structure of Nup120 | | Descriptor: | MERCURY (II) ION, Nucleoporin NUP120 | | Authors: | Seo, H.S, Ma, Y, Debler, E.W, Blobel, G, Hoelz, A. | | Deposit date: | 2008-11-08 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3H7N
 
 | | Structure of Nup120 | | Descriptor: | Nucleoporin NUP120 | | Authors: | Seo, H.S, Ma, Y, Debler, E.W, Blobel, G, Hoelz, A. | | Deposit date: | 2009-04-27 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5ZBJ
 
 | |
6A6Q
 
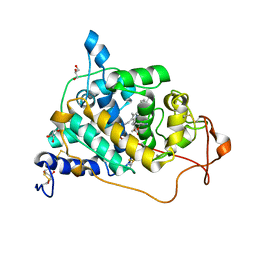 | | Crystal structure of a lignin peroxidase isozyme H8 variant that is stable at very acidic pH | | Descriptor: | CALCIUM ION, GLYCEROL, HEME B/C, ... | | Authors: | Seo, H, Kim, K.-J, Pham, L.T.M. | | Deposit date: | 2018-06-29 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | In silico-designed lignin peroxidase fromPhanerochaete chrysosporiumshows enhanced acid stability for depolymerization of lignin.
Biotechnol Biofuels, 11, 2018
|
|
5ZI9
 
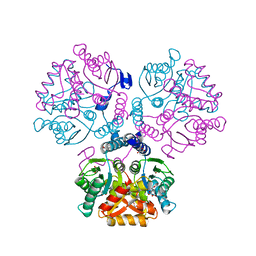 | | Crystal structure of type-II LOG from Streptomyces coelicolor A3 | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Cytokinin riboside 5'-monophosphate phosphoribohydrolase, ... | | Authors: | Seo, H, Kim, K.-J. | | Deposit date: | 2018-03-14 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical characterization of the type-II LOG protein from Streptomyces coelicolor A3.
Biochem. Biophys. Res. Commun., 499, 2018
|
|
5ZBK
 
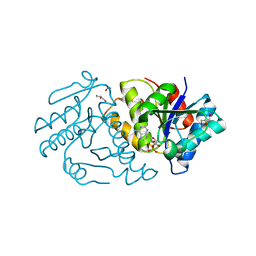 | |
5ZBL
 
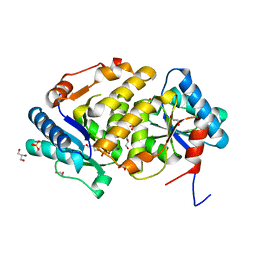 | |
8CUC
 
 | |
8ECM
 
 | |
4MHC
 
 | | Crystal Structure of a Nucleoporin | | Descriptor: | Nucleoporin NUP157 | | Authors: | Seo, H.S, Blus, B.J, Blobel, G. | | Deposit date: | 2013-08-29 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and nucleic acid binding activity of the nucleoporin Nup157.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5WQ3
 
 | |
7DIB
 
 | |
7DG5
 
 | |
6UCH
 
 | | SMARCB1 nucleosome-interacting C-terminal alpha helix | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Valencia, A.M, Sun, Z.Y.J, Seo, H.S, Vangos, H.S, Yeoh, Z.C, Mashtalir, N, Dhe-Paganon, S, Kadoch, C. | | Deposit date: | 2019-09-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recurrent SMARCB1 Mutations Reveal a Nucleosome Acidic Patch Interaction Site That Potentiates mSWI/SNF Complex Chromatin Remodeling.
Cell, 179, 2019
|
|
4OWR
 
 | | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair Rae1-Nup98 | | Descriptor: | Matrix protein, Nuclear pore complex protein Nup98-Nup96, mRNA export factor | | Authors: | Ren, Y, Quan, B, Seo, H.S, Blobel, G. | | Deposit date: | 2014-02-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 Nup98).
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6J57
 
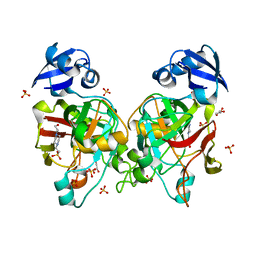 | | Crystal structure of fumarylpyruvate hydrolase from Corynebacterium glutamicum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Predicted 2-keto-4-pentenoate hydratase/2-oxohepta-3-ene-1,7-dioic acid hydratase, ... | | Authors: | Hong, H, Seo, H, Kim, K.-J, Park, W. | | Deposit date: | 2019-01-10 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Sequence, structure and function-based classification of the broadly conserved FAH superfamily reveals two distinct fumarylpyruvate hydrolase subfamilies.
Environ.Microbiol., 22, 2020
|
|
6J5Y
 
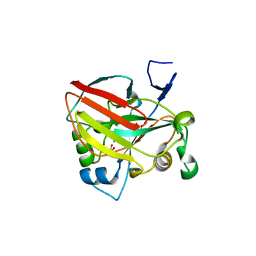 | | Crystal structure of fumarylpyruvate hydrolase from Pseudomonas aeruginosa in complex with Mn2+ and pyruvate | | Descriptor: | FAA hydrolase family protein, MANGANESE (II) ION, PYRUVIC ACID | | Authors: | Hong, H, Seo, H, Kim, K.-J, Park, W. | | Deposit date: | 2019-01-12 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Sequence, structure and function-based classification of the broadly conserved FAH superfamily reveals two distinct fumarylpyruvate hydrolase subfamilies.
Environ.Microbiol., 22, 2020
|
|
8HKA
 
 | |
8HK9
 
 | |
8HKB
 
 | |
