6KNF
 
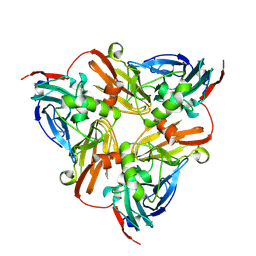 | | CryoEM map and model of Nitrite Reductase at pH 6.2 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Adachi, N, Yamaguchi, T, Moriya, T, Kawasaki, M, Koiwai, K, Shinoda, A, Yamada, Y, Yumoto, F, Kohzuma, T, Senda, T. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | 2.85 and 2.99 angstrom resolution structures of 110 kDa nitrite reductase determined by 200 kV cryogenic electron microscopy.
J.Struct.Biol., 213, 2021
|
|
7DNM
 
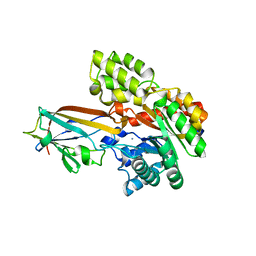 | | Crystal structure of the AgCarB2-C2 complex | | Descriptor: | AP_endonuc_2 domain-containing protein, AgCarC2, IODIDE ION, ... | | Authors: | Senda, M, Kumano, T, Watanabe, S, Kobayashi, M, Senda, T. | | Deposit date: | 2020-12-10 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the metabolism of xenobiotic C-glycosides by intestinal bacteria
Nat Commun, 2021
|
|
6KNG
 
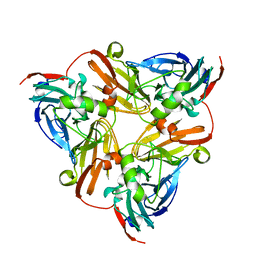 | | CryoEM map and model of Nitrite Reductase at pH 8.1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Adachi, N, Yamaguchi, T, Moriya, T, Kawasaki, M, Koiwai, K, Shinoda, A, Yamada, Y, Yumoto, F, Kohzuma, T, Senda, T. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | 2.85 and 2.99 angstrom resolution structures of 110 kDa nitrite reductase determined by 200 kV cryogenic electron microscopy.
J.Struct.Biol., 213, 2021
|
|
1KJO
 
 | |
1KRO
 
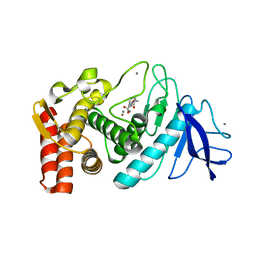 | |
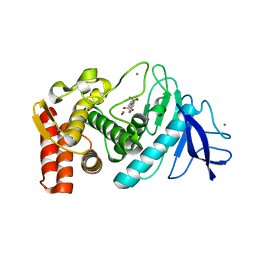
1KJP
 
 | |
7DRD
 
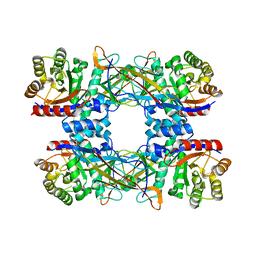 | | Cryo-EM structure of DgpB-C at 2.85 angstrom resolution | | Descriptor: | AP_endonuc_2 domain-containing protein, DgpB | | Authors: | Mori, T, Moriya, T, Adachi, N, Senda, T, Abe, I. | | Deposit date: | 2020-12-28 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7DRE
 
 | | Cryo-EM structure of DfgA-B at 2.54 angstrom resolution | | Descriptor: | DfgB, Sugar phosphate isomerase/epimerase | | Authors: | Mori, T, Moriya, T, Adachi, N, Senda, T, Abe, I. | | Deposit date: | 2020-12-28 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
2IO5
 
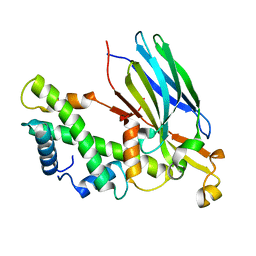 | | Crystal structure of the CIA- histone H3-H4 complex | | Descriptor: | ASF1A protein, Histone H3.1, Histone H4 | | Authors: | Natsume, R, Akai, Y, Horikoshi, M, Senda, T. | | Deposit date: | 2006-10-10 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4.
Nature, 446, 2007
|
|
1C4X
 
 | | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE (BPHD) FROM RHODOCOCCUS SP. STRAIN RHA1 | | Descriptor: | PROTEIN (2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE) | | Authors: | Nandhagopal, N, Senda, T, Mitsui, Y. | | Deposit date: | 1999-09-30 | | Release date: | 1999-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-Dimensional Structure of Microbial 2-Hydroxyl-6-Oxo-6-Phenylhexa-2,4- Dienoic Acid (Hpda) Hydrolase (Bphd Enzyme) from Rhodococcus Sp. Strain Rha1, in the Pcb Degradation Pathway
Proc.Jpn.Acad.,Ser.B, 73, 1997
|
|
6KEM
 
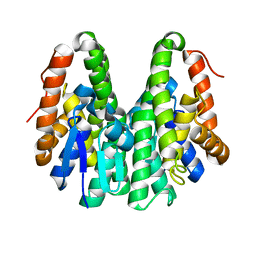 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in apo-form 2 | | Descriptor: | Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Niwa, R, Senda, T. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
6KEL
 
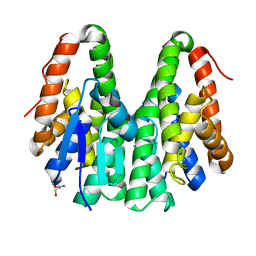 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in apo-form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
6KEQ
 
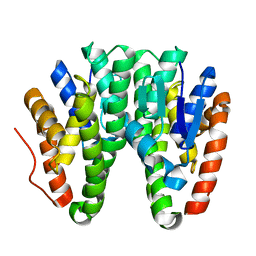 | | Crystal structure of D113A mutant of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in apo-form | | Descriptor: | Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Niwa, R, Senda, T. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
6KER
 
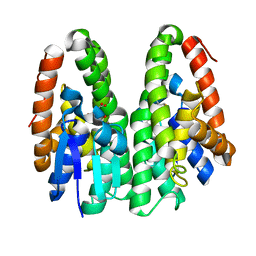 | | Crystal structure of D113A mutant of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in glutathione-bound form | | Descriptor: | GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Niwa, R, Senda, T. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
6KEN
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in glutathione-bound form | | Descriptor: | GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Niwa, R, Senda, T. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
6KEO
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in 17beta-estradiol-bound form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ESTRADIOL, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Niwa, R, Senda, T. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
6KEP
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in 17beta-estradiol- and glutathione-bound form | | Descriptor: | ESTRADIOL, GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Niwa, R, Senda, T. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
3WRC
 
 | | Crystal structure of the anaerobic DesB-Protocatechuate (PCA) complex | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (II) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2014-02-21 | | Release date: | 2014-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
3WR9
 
 | | Crystal structure of the Anaerobic DesB-Gallate complex | | Descriptor: | 3,4,5-trihydroxybenzoic acid, FE (II) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2014-02-21 | | Release date: | 2014-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
3WRA
 
 | | Crystal structure of the desB-Gallate complex exposed to Aerobic Atomosphere | | Descriptor: | FE (II) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2014-02-21 | | Release date: | 2014-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
3WR3
 
 | | Crystal structure of the anaerobic DesB-gallate complex | | Descriptor: | 3,4,5-trihydroxybenzoic acid, FE (II) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
3WKU
 
 | | Crystal structure of the anaerobic DesB-gallate complex | | Descriptor: | 3,4,5-trihydroxybenzoic acid, FE (III) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2013-10-31 | | Release date: | 2014-04-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
3WR8
 
 | | Crystal structure of DesB from Sphingobium sp. strain SYK-6 | | Descriptor: | FE (II) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2014-02-21 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
3WPM
 
 | | Crystal structure of the anaerobic DesB-gallate complex by co-crystallization | | Descriptor: | 3,4,5-trihydroxybenzoic acid, FE (III) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2014-01-14 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
3WR4
 
 | | Crystal structure of the anaerobic DesB-gallate complex | | Descriptor: | 3,4,5-trihydroxybenzoic acid, FE (III) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
