4DY3
 
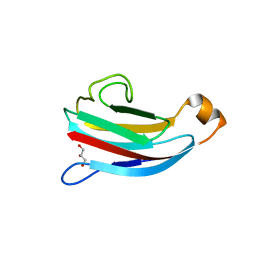 | | crystal structure of Escherichia coli PliG, a periplasmic lysozyme inhibitor of g-type lysozyme | | Descriptor: | GLYCEROL, Inhibitor of g-type lysozyme | | Authors: | Leysen, S, Vanheuverzwijn, S, Van Asten, K, Vanderkelen, L, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2012-02-28 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the PliG lysozyme inhibitor family.
J.Struct.Biol., 180, 2012
|
|
8CEY
 
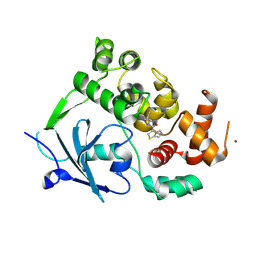 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH11233. | | Descriptor: | N-glycosylase/DNA lyase, NICKEL (II) ION, ~{N}-(3,3-dimethylcyclobutyl)imidazo[5,1-b][1,3]thiazole-3-carboxamide | | Authors: | Kosenina, S, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Virtual fragment screening for DNA repair inhibitors in vast chemical space.
Nat Commun, 16, 2025
|
|
8CEX
 
 | |
4ID6
 
 | |
6DD8
 
 | | Structure of mouse SYCP3, P21 form | | Descriptor: | Synaptonemal complex protein 3 | | Authors: | Rosenberg, S.C, Munoz, I.C, Uson, I, Corbett, K.D. | | Deposit date: | 2018-05-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A conserved filamentous assembly underlies the structure of the meiotic chromosome axis.
Elife, 8, 2019
|
|
6NYA
 
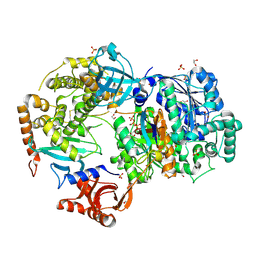 | | Crystal Structure of ubiquitin E1 (Uba1) in complex with Ubc3 (Cdc34) and ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Olsen, S.K, Williams, K.M, Atkison, J.H. | | Deposit date: | 2019-02-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.065 Å) | | Cite: | Structural insights into E1 recognition and the ubiquitin-conjugating activity of the E2 enzyme Cdc34.
Nat Commun, 10, 2019
|
|
6DD9
 
 | | Structure of mouse SYCP3, P1 form | | Descriptor: | Synaptonemal complex protein 3 | | Authors: | Rosenberg, S.C, Munoz, I.C, Uson, I, Corbett, K.D. | | Deposit date: | 2018-05-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A conserved filamentous assembly underlies the structure of the meiotic chromosome axis.
Elife, 8, 2019
|
|
6NYD
 
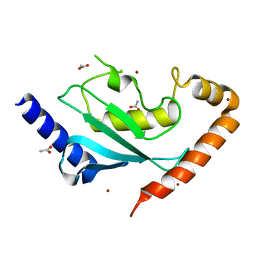 | |
3SPC
 
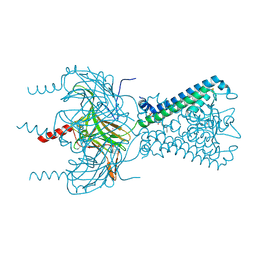 | | Inward rectifier potassium channel Kir2.2 in complex with dioctanoylglycerol pyrophosphate (DGPP) | | Descriptor: | (2R)-3-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}propane-1,2-diyl dioctanoate, Inward-rectifier K+ channel Kir2.2, POTASSIUM ION | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.454 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
3SPG
 
 | | Inward rectifier potassium channel Kir2.2 R186A mutant in complex with PIP2 | | Descriptor: | Inward-rectifier K+ channel Kir2.2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
5MA3
 
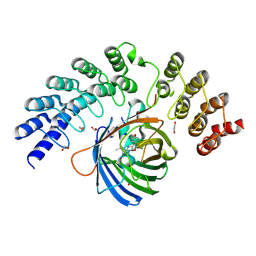 | | GFP-binding DARPin fusion gc_R11 | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, R11 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
6O82
 
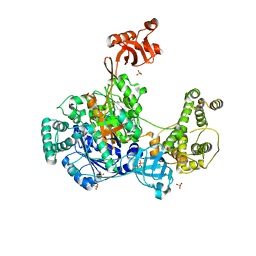 | | S. pombe ubiquitin E1 complex with a ubiquitin-AMP mimic | | Descriptor: | 1,2-ETHANEDIOL, 5'-deoxy-5'-(sulfamoylamino)adenosine, MAGNESIUM ION, ... | | Authors: | Olsen, S.K, Lima, C.D. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structural basis for adenylation and thioester bond formation in the ubiquitin E1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5MFD
 
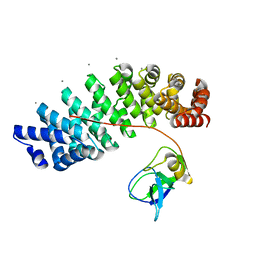 | | Designed armadillo repeat protein YIIIM''6AII in complex with pD_(KR)5 | | Descriptor: | CALCIUM ION, Capsid decoration protein,pD_(KR)5, YIIIM''6AII | | Authors: | Hansen, S, Kiefer, J, Madhurantakam, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of designed armadillo repeat proteins binding to peptides fused to globular domains.
Protein Sci., 26, 2017
|
|
5MFJ
 
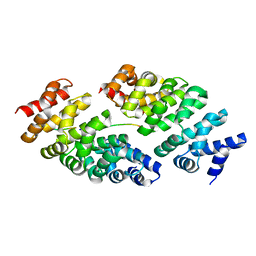 | | Designed armadillo repeat protein YIII(Dq.V2)4CqI in complex with peptide (KR)5 | | Descriptor: | (KR)5, YIII(Dq.V2)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFI
 
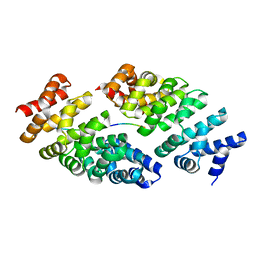 | | Designed armadillo repeat protein YIII(Dq.V2)4CqI in complex with peptide (KR)4 | | Descriptor: | (KR)4, YIII(Dq.V2)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFK
 
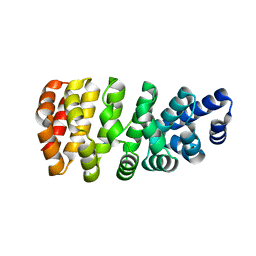 | | Designed armadillo repeat protein YIII(Dq.V1)4CPAF in complex with peptide (KR)4 | | Descriptor: | (KR)4, YIII(Dq.V1)4CPAF | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
3SPJ
 
 | |
3P0G
 
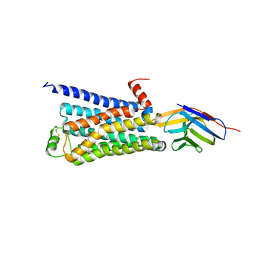 | | Structure of a nanobody-stabilized active state of the beta2 adrenoceptor | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Beta-2 adrenergic receptor, Lysozyme, ... | | Authors: | Rasmussen, S.G.F, Choi, H.-J, Fung, J.J, Pardon, E, Casarosa, P, Chae, P.S, DeVree, B.T, Rosenbaum, D.M, Thian, F.S, Kobilka, T.S, Schnapp, A, Konetzki, I, Sunahara, R.K, Gellman, S.H, Pautsch, A, Steyaert, J, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2010-09-28 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a nanobody-stabilized active state of the b2 adrenoceptor
Nature, 469, 2011
|
|
4QMS
 
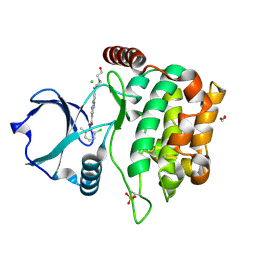 | | MST3 in complex with DASATINIB | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, ... | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
4QO9
 
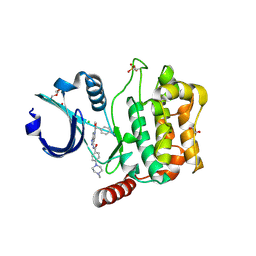 | | MST3 IN COMPLEX WITH Danusertib | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
6ZPN
 
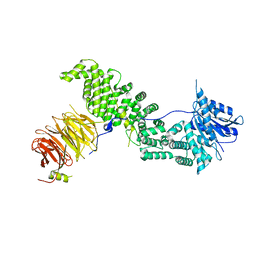 | | Crystal structure of Chaetomium thermophilum Raptor | | Descriptor: | WD_REPEATS_REGION domain-containing protein | | Authors: | Imseng, S, Boehm, R, Jakob, R.P, Hall, M.N, Hiller, S, Maier, T. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The dynamic mechanism of 4E-BP1 recognition and phosphorylation by mTORC1.
Mol.Cell, 81, 2021
|
|
5MFN
 
 | | Designed armadillo repeat protein YIIIM5AII | | Descriptor: | CALCIUM ION, D-MALATE, YIIIM5AII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFB
 
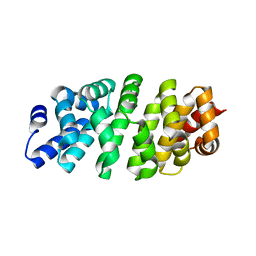 | | Designed armadillo repeat protein YIII(Dq)4CqI | | Descriptor: | YIII(Dq)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MA5
 
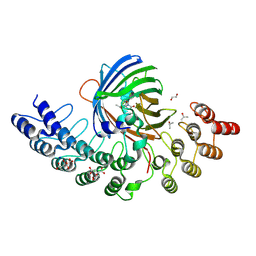 | | GFP-binding DARPin fusion gc_K11 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Green fluorescent protein, ... | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MAD
 
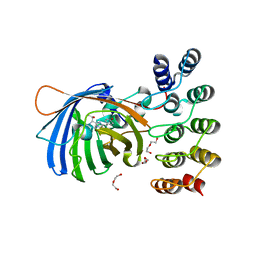 | | GFP-binding DARPin 3G61 | | Descriptor: | 3G61, DI(HYDROXYETHYL)ETHER, Green fluorescent protein, ... | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
