8UFJ
 
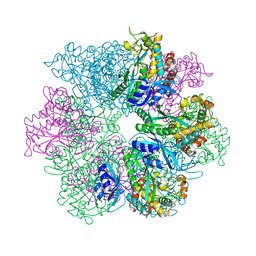 | | Structure of M. mazei GS(R167L-A168G) apo form | | Descriptor: | Glutamine synthetase, MAGNESIUM ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-10-04 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | M. mazei glutamine synthetase and glutamine synthetase-GlnK1 structures reveal enzyme regulation by oligomer modulation.
Nat Commun, 14, 2023
|
|
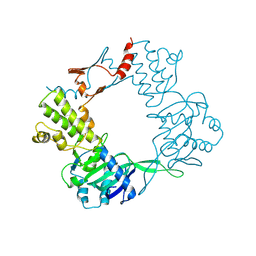
9BE2
 
 | |
1ZVV
 
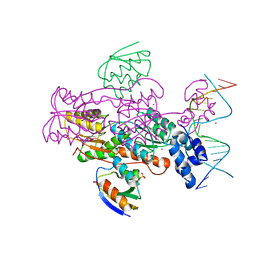 | | Crystal structure of a ccpa-crh-dna complex | | Descriptor: | DNA recognition strand CRE, Glucose-resistance amylase regulator, HPr-like protein crh, ... | | Authors: | Schumacher, M.A, Brennan, R.G, Hillen, W, Seidel, G. | | Deposit date: | 2005-06-02 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Phosphoprotein Crh-Ser46-P displays altered binding to CcpA to effect carbon catabolite regulation.
J.Biol.Chem., 281, 2006
|
|
6E4N
 
 | |
6E4O
 
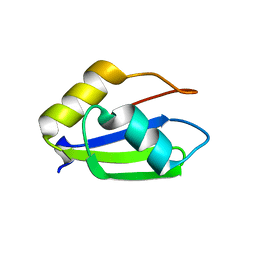 | | Structure of apo T. brucei RRM: P4(1)2(1)2 form | | Descriptor: | RNA-binding protein, putative | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-07-18 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The RRM of the kRNA-editing protein TbRGG2 uses multiple surfaces to bind and remodel RNA.
Nucleic Acids Res., 47, 2019
|
|
6E4P
 
 | | Structure of the T. brucei RRM domain in complex with RNA | | Descriptor: | RNA (5'-R(P*UP*UP*UP*U)-3'), RNA-binding protein, putative | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-07-18 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | The RRM of the kRNA-editing protein TbRGG2 uses multiple surfaces to bind and remodel RNA.
Nucleic Acids Res., 47, 2019
|
|
6ALX
 
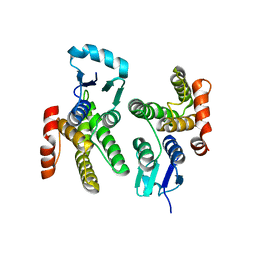 | |
6AMK
 
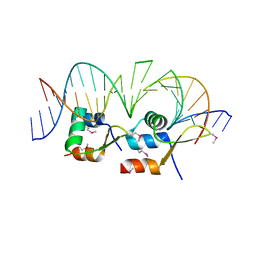 | | Structure of Streptomyces venezuelae BldC-whiI opt complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*CP*CP*GP*AP*AP*TP*TP*AP*CP*CP*CP*GP*AP*AP*TP*TP*G)-3'), DNA (5'-D(*TP*TP*CP*AP*AP*TP*TP*CP*GP*GP*GP*TP*AP*AP*TP*TP*CP*GP*GP*GP*CP*A)-3'), Putative DNA-binding protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2017-08-09 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.288 Å) | | Cite: | The MerR-like protein BldC binds DNA direct repeats as cooperative multimers to regulate Streptomyces development.
Nat Commun, 9, 2018
|
|
6AMA
 
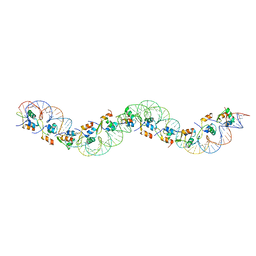 | |
3BTI
 
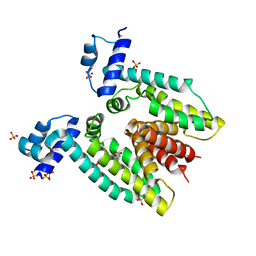 | |
3BTC
 
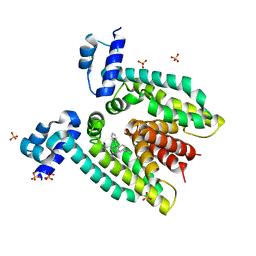 | |
3BTJ
 
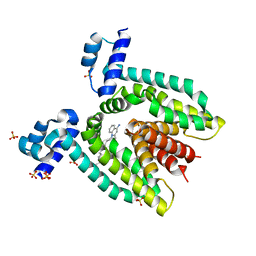 | |
3BTL
 
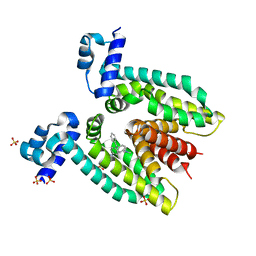 | |
7RMW
 
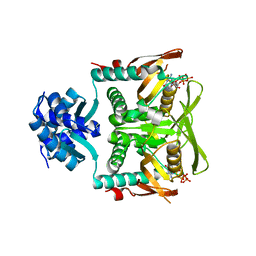 | | Crystal structure of B. subtilis PurR bound to ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, Pur operon repressor | | Authors: | Schumacher, M.A. | | Deposit date: | 2021-07-28 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The nucleotide messenger (p)ppGpp is an anti-inducer of the purine synthesis transcription regulator PurR in Bacillus.
Nucleic Acids Res., 50, 2022
|
|
8DPK
 
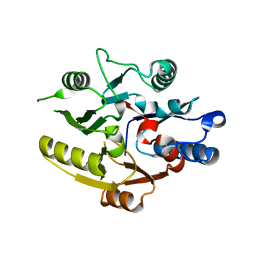 | |
1QVT
 
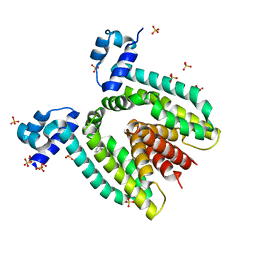 | |
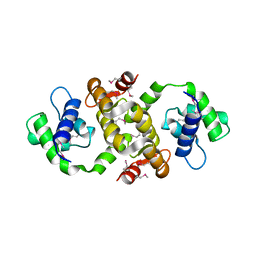
1QX7
 
 | |
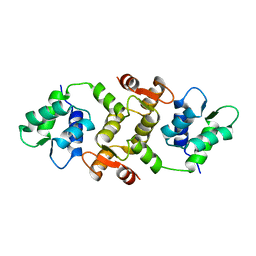
1QX5
 
 | |
8CSH
 
 | |
6UEP
 
 | |
6U9X
 
 | | Structure of T. brucei MERS1-RNA complex | | Descriptor: | Mitochondrial edited mRNA stability factor 1, RNA (5'-R(*GP*AP*GP*AP*GP*GP*GP*GP*GP*UP*U)-3') | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-09-09 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of MERS1, the 5' processing enzyme of mitochondrial mRNAs inTrypanosoma brucei.
Rna, 26, 2020
|
|
6UEO
 
 | | Structure of A. thaliana TBP-AC mismatch DNA site | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*CP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TATA-box-binding protein 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA mismatches reveal conformational penalties in protein-DNA recognition.
Nature, 587, 2020
|
|
6UER
 
 | |
6UMK
 
 | | Structure of E. coli FtsZ(L178E)-GDP complex | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution crystal structures of Escherichia coli FtsZ bound to GDP and GTP.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4RX6
 
 | | Structure of B. subtilis GlnK-ATP complex to 2.6 Angstrom | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Nitrogen regulatory PII-like protein | | Authors: | Schumacher, M.A, Cuthbert, B, Tonthat, N, Chinnam, N.G, Whitfill, T. | | Deposit date: | 2014-12-09 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5994 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
