1H3C
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, N-(6-{[3-(4-BROMOPHENYL)-1,2-BENZISOTHIAZOL-6-YL]OXY}HEXYL)-N-METHYLPROP-2-EN-1-AMINE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-25 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1GSZ
 
 | | Crystal Structure of a Squalene Cyclase in Complex with the Potential Anticholesteremic Drug Ro48-8071 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE, [4-({6-[ALLYL(METHYL)AMINO]HEXYL}OXY)-2-FLUOROPHENYL](4-BROMOPHENYL)METHANONE | | Authors: | Lenhart, A, Weihofen, W.A, Pleschke, A.E.W, Schulz, G.E. | | Deposit date: | 2002-01-09 | | Release date: | 2003-01-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Squalene Cyclase in Complex with the Potential Anticholesteremic Drug Ro48-8071
Chem.Biol., 9, 2002
|
|
1H35
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (4'-{[ALLYL(METHYL)AMINO]METHYL}-1,1'-BIPHENYL-4-YL)(4-BROMOPHENYL)METHANONE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H37
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE, {4-[((1S,2S)-2-{[ALLYL(CYCLOPROPYL)AMINO]METHYL}CYCLOPROPYL)METHOXY]PHENYL}(4-BROMOPHENYL)METHANONE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H36
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (4-BROMOPHENYL)[4-({(2E)-4-[CYCLOPROPYL(METHYL)AMINO]BUT-2-ENYL}OXY)PHENYL]METHANONE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H39
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, ALLYL-{6-[3-(4-BROMO-PHENYL)-1-METHYL-1H-INDAZOL-6-YL]OXY}HEXYL)-N-METHYLAMINE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
8CGT
 
 | | STRUCTURE OF CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED WITH A THIO-MALTOHEXAOSE | | Descriptor: | CALCIUM ION, PROTEIN (CYCLODEXTRIN-GLYCOSYLTRANSFERASE), alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose | | Authors: | Schmidt, A.K, Schulz, G.E. | | Deposit date: | 1998-09-27 | | Release date: | 1998-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate binding to a cyclodextrin glycosyltransferase and mutations increasing the gamma-cyclodextrin production.
Eur.J.Biochem., 255, 1998
|
|
1FUA
 
 | | L-FUCULOSE 1-PHOSPHATE ALDOLASE CRYSTAL FORM T | | Descriptor: | BETA-MERCAPTOETHANOL, L-FUCULOSE-1-PHOSPHATE ALDOLASE, SULFATE ION, ... | | Authors: | Dreyer, M.K, Schulz, G.E. | | Deposit date: | 1996-02-14 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Refined high-resolution structure of the metal-ion dependent L-fuculose-1-phosphate aldolase (class II) from Escherichia coli.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1UMP
 
 | |
1MPR
 
 | | MALTOPORIN FROM SALMONELLA TYPHIMURIUM | | Descriptor: | CALCIUM ION, MALTOPORIN | | Authors: | Meyer, J.E.W, Schulz, G.E. | | Deposit date: | 1996-12-18 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of maltoporin from Salmonella typhimurium ligated with a nitrophenyl-maltotrioside.
J.Mol.Biol., 266, 1997
|
|
1NKS
 
 | |
1O9G
 
 | |
3FUA
 
 | | L-FUCULOSE-1-PHOSPHATE ALDOLASE CRYSTAL FORM K | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, L-FUCULOSE-1-PHOSPHATE ALDOLASE, ... | | Authors: | Dreyer, M.K, Schulz, G.E. | | Deposit date: | 1996-02-14 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Catalytic mechanism of the metal-dependent fuculose aldolase from Escherichia coli as derived from the structure.
J.Mol.Biol., 259, 1996
|
|
1GQC
 
 | |
1GQ9
 
 | |
1GET
 
 | | ANATOMY OF AN ENGINEERED NAD-BINDING SITE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mittl, P.R.E, Schulz, G.E. | | Deposit date: | 1994-01-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anatomy of an engineered NAD-binding site.
Protein Sci., 3, 1994
|
|
1GK2
 
 | |
1GK3
 
 | |
1GKY
 
 | |
1GES
 
 | |
1GKM
 
 | |
1GEU
 
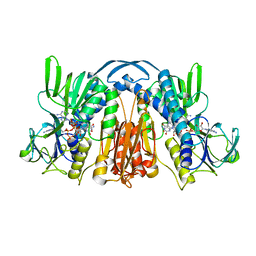 | | ANATOMY OF AN ENGINEERED NAD-BINDING SITE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mittl, P.R.E, Schulz, G.E. | | Deposit date: | 1994-01-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anatomy of an engineered NAD-binding site.
Protein Sci., 3, 1994
|
|
2VSA
 
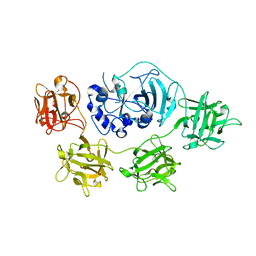 | | Structure and mode of action of a mosquitocidal holotoxin | | Descriptor: | MOSQUITOCIDAL TOXIN | | Authors: | Treiber, N, Reinert, D.J, Carpusca, I, Aktories, K, Schulz, G.E. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure and Mode of Action of a Mosquitocidal Holotoxin
J.Mol.Biol., 381, 2008
|
|
2VSE
 
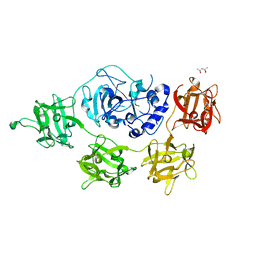 | | Structure and mode of action of a mosquitocidal holotoxin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, MOSQUITOCIDAL TOXIN | | Authors: | Treiber, N, Reinert, D.J, Carpusca, I, Aktories, K, Schulz, G.E. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mode of Action of a Mosquitocidal Holotoxin.
J.Mol.Biol., 381, 2008
|
|
3E1T
 
 | | Structure and action of the myxobacterial chondrochloren halogenase CndH, a new variant of FAD-dependent halogenases | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Halogenase | | Authors: | Buedenbender, S, Rachid, S, Mueller, R, Schulz, G.E. | | Deposit date: | 2008-08-04 | | Release date: | 2009-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and action of the myxobacterial chondrochloren halogenase CndH: a new variant of FAD-dependent halogenases.
J.Mol.Biol., 385, 2009
|
|
