1SXI
 
 | | Structure of apo transcription regulator B. megaterium | | Descriptor: | Glucose-resistance amylase regulator, MAGNESIUM ION | | Authors: | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
1SXG
 
 | | Structural studies on the apo transcription factor form B. megaterium | | Descriptor: | 2-PHENYLAMINO-ETHANESULFONIC ACID, Glucose-resistance amylase regulator | | Authors: | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
4RS7
 
 | |
3FBR
 
 | | structure of HipA-amppnp-peptide | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Serine/threonine-protein kinase toxin HipA, peptide of EF-Tu | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-11-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
1QVU
 
 | |
4GCK
 
 | | structure of no-dna complex | | Descriptor: | DNA (5'-D(*GP*TP*GP*AP*GP*TP*AP*CP*TP*CP*AP*C)-3'), Nucleoid occlusion factor SlmA | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-07-30 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GFL
 
 | | NO mechanism, slma | | Descriptor: | Nucleoid occlusion factor SlmA | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-08-03 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1SN8
 
 | | Crystal structure of the S1 domain of RNase E from E. coli (Pb derivative) | | Descriptor: | LEAD (II) ION, Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
1SMX
 
 | | Crystal structure of the S1 domain of RNase E from E. coli (native) | | Descriptor: | Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
1RZR
 
 | | crystal structure of transcriptional regulator-phosphoprotein-DNA complex | | Descriptor: | 5'-D(*CP*TP*GP*AP*AP*AP*GP*CP*GP*CP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*AP*GP*CP*GP*CP*TP*TP*TP*CP*AP*G)-3', Glucose-resistance amylase regulator, ... | | Authors: | Schumacher, M.A, Allen, G.S, Brennan, R.G. | | Deposit date: | 2003-12-27 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P.
Cell(Cambridge,Mass.), 118, 2004
|
|
1KQ2
 
 | | Crystal Structure of an Hfq-RNA Complex | | Descriptor: | 5'-R(*AP*UP*UP*UP*UP*UP*G)-3', Host factor for Q beta | | Authors: | Schumacher, M.A, Pearson, R.F, Moller, T, Valentin-Hansen, P, Brennan, R.G. | | Deposit date: | 2002-01-03 | | Release date: | 2002-07-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein.
EMBO J., 21, 2002
|
|
1SXH
 
 | | apo structure of B. megaterium transcription regulator | | Descriptor: | Glucose-resistance amylase regulator | | Authors: | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
4R25
 
 | | Structure of B. subtilis GlnK | | Descriptor: | Nitrogen regulatory PII-like protein, ZINC ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5193 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
3EZ2
 
 | | Partition protein-ADP complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Schumacher, M.A, Dunham, T.D, Xu, W, Funnell, B. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA.
Embo J., 28, 2009
|
|
1YM8
 
 | |
1SLJ
 
 | | Solution structure of the S1 domain of RNase E from E. coli | | Descriptor: | Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-05 | | Release date: | 2004-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
5QHZ
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000385a | | Descriptor: | 2-cyano-~{N}-cyclohexyl-ethanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QHV
 
 | | PanDDA analysis group deposition -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000299a | | Descriptor: | 1-cyclohexyl-3-(2-pyridin-4-ylethyl)urea, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QHY
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000462a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI7
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000506a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QHU
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMSOA000341b | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, N-(2-hydroxyphenyl)acetamide, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI4
 
 | | PanDDA analysis group deposition -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000466a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QI0
 
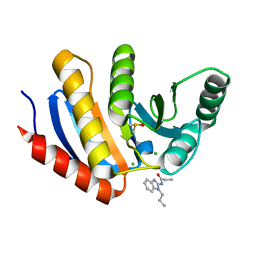 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000352a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QHW
 
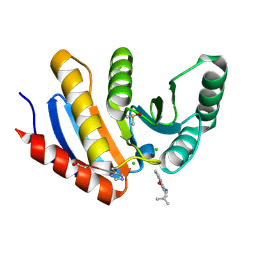 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000347a | | Descriptor: | 2-methyl-~{N}-(2-methylpropyl)imidazo[1,2-a]pyridine-3-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI3
 
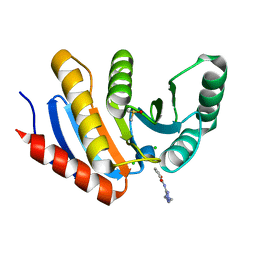 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000475a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
