1DYM
 
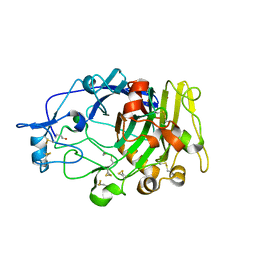 | | Humicola insolens Endocellulase Cel7B (EG 1) E197A Mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Moraz, O, Driguez, H, Schulein, M. | | Deposit date: | 2000-02-03 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate.
Biochem.J., 335 ( Pt 2), 1998
|
|
1DXI
 
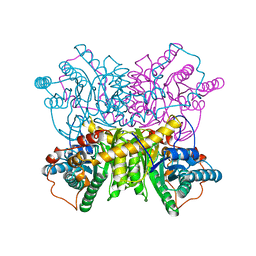 | | STRUCTURE DETERMINATION OF GLUCOSE ISOMERASE FROM STREPTOMYCES MURINUS AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION | | Authors: | Rasmussen, H, La Cour, T, Nyborg, J, Schulein, M. | | Deposit date: | 1993-09-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure determination of glucose isomerase from Streptomyces murinus at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1OCJ
 
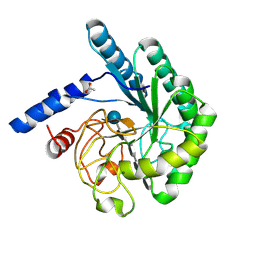 | | Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with a THIOPENTASACCHARIDE at 1.3 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-07 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
1OC5
 
 | | D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with methyl-cellobiosyl-4-deoxy-4-thio-beta-D-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, GLYCEROL, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-06 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
1OC7
 
 | | D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with methyl-tetrathio-alpha-d-cellopentoside at 1.1 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-06 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
1M8M
 
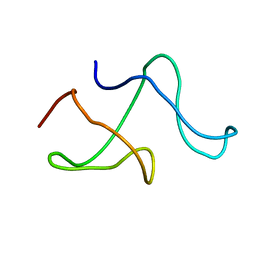 | | SOLID-STATE MAS NMR STRUCTURE OF THE A-SPECTRIN SH3 DOMAIN | | Descriptor: | SPECTRIN ALPHA CHAIN, BRAIN | | Authors: | Castellani, F, Van Rossum, B, Diehl, A, Schubert, M, Rehbein, K, Oschkinat, H. | | Deposit date: | 2002-07-25 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy
Nature, 420, 2002
|
|
1OC6
 
 | | structure native of the D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS at 1.5 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-06 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
1QI2
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE TETRAGONAL CRYSTAL FORM IN COMPLEX WITH 2',4'-DINITROPHENYL 2-DEOXY-2-FLUORO-B-D-CELLOTRIOSIDE | | Descriptor: | CALCIUM ION, ENDOGLUCANASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Varrot, A, Schulein, M, Davies, G.J. | | Deposit date: | 1999-06-02 | | Release date: | 2000-06-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into ligand-induced conformational change in Cel5A from Bacillus agaradhaerens revealed by a catalytically active crystal form.
J.Mol.Biol., 297, 2000
|
|
1QI0
 
 | |
5U56
 
 | | Structure of Francisella tularensis heterodimeric SspA (MglA-SspA) | | Descriptor: | GLYCEROL, Macrophage growth locus A, PENTAETHYLENE GLYCOL, ... | | Authors: | Cuthbert, B.J, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2016-12-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dissection of the molecular circuitry controlling virulence in Francisella tularensis.
Genes Dev., 31, 2017
|
|
5U51
 
 | | Structure of Francisella tularensis heterodimeric SspA (MglA-SspA) in complex with ppGpp | | Descriptor: | GLYCEROL, GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Cuthbert, B.J, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2016-12-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dissection of the molecular circuitry controlling virulence in Francisella tularensis.
Genes Dev., 31, 2017
|
|
1QHZ
 
 | |
1E5J
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE TETRAGONAL CRYSTAL FORM IN COMPLEX WITH METHYL-4II-S-ALPHA-CELLOBIOSYL-4II-THIO-BETA-CELLOBIOSIDE | | Descriptor: | CALCIUM ION, ENDOGLUCANASE 5A, alpha-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside | | Authors: | Fort, S, Varrot, A, Schulein, M, Cottaz, S, Driguez, H, Davies, G.J. | | Deposit date: | 2000-07-26 | | Release date: | 2001-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mixed-Linkage Cellooligosaccharides: A New Class of Glycoside Hydrolase Inhibitors
Chembiochem, 2, 2001
|
|
1BVW
 
 | | CELLOBIOHYDROLASE II (CEL6A) FROM HUMICOLA INSOLENS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Varrot, A, Davies, G.J, Schulein, M. | | Deposit date: | 1998-09-19 | | Release date: | 1999-09-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of the catalytic core domain of the family 6 cellobiohydrolase II, Cel6A, from Humicola insolens, at 1.92 A resolution.
Biochem.J., 337, 1999
|
|
1H4H
 
 | | Oligosaccharide-binding to family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre | | Descriptor: | XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Sabini, E, Wilson, K.S, Danielsen, S, Schulein, M, Davies, G.J. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalysis and Specificity in Enzymatic Glycoside Hydrolysis: A 2,5B Conformation for the Glycosyl-Enzyme Intermediate Revealed by the Structure of the Bacillus Agaradhaerens Family 11 Xylanase.
Chem.Biol., 6, 1999
|
|
1I9B
 
 | | X-RAY STRUCTURE OF ACETYLCHOLINE BINDING PROTEIN (ACHBP) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYLCHOLINE BINDING PROTEIN, CALCIUM ION | | Authors: | Brejc, K, van Dijk, W.J, Klaassen, R, Schuurmans, M, van der Oost, J, Smit, A.B, Sixma, T.K. | | Deposit date: | 2001-03-18 | | Release date: | 2001-05-16 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors.
Nature, 411, 2001
|
|
1H4G
 
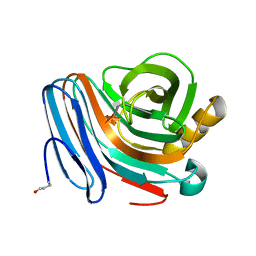 | | Oligosaccharide-binding to family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre | | Descriptor: | SULFATE ION, XYLANASE, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Sabini, E, Wilson, K.S, Danielsen, S, Schulein, M, Davies, G.J. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Catalysis and Specificity in Enzymatic Glycoside Hydrolysis: A 2,5B Conformation for the Glycosyl-Enzyme Intermediate Revealed by the Structure of the Bacillus Agaradhaerens Family 11 Xylanase.
Chem.Biol., 6, 1999
|
|
1H5V
 
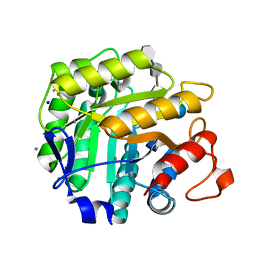 | | Thiopentasaccharide complex of the endoglucanase Cel5A from Bacillus agaradharens at 1.1 A resolution in the tetragonal crystal form | | Descriptor: | CALCIUM ION, ENDOGLUCANASE 5A, SODIUM ION, ... | | Authors: | Varrot, A, Sulzenbacher, G, Schulein, M, Driguez, H, Davies, G.J. | | Deposit date: | 2001-05-28 | | Release date: | 2002-05-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structure of Endoglucanase Cel5A in Complex with Methyl 4,4II,4III,4Iv-Tetrathio-Alpha-Cellopentoside Highlights the Alternative Binding Modes Targeted by Substrate Mimics
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5S4J
 
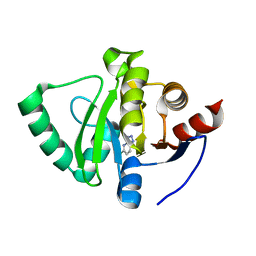 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF054 | | Descriptor: | 6-chlorotetrazolo[1,5-b]pyridazine, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.124 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4I
 
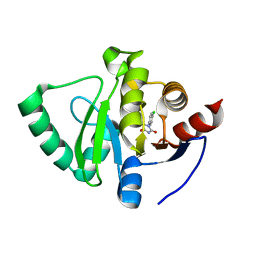 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF051 | | Descriptor: | (5S)-1-(4-chlorophenyl)-5-methylimidazolidine-2,4-dione, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S18
 
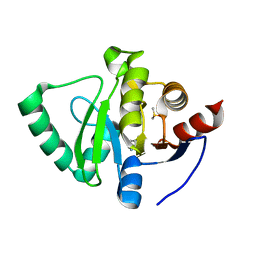 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with EN300-321461 | | Descriptor: | 6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidine, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S20
 
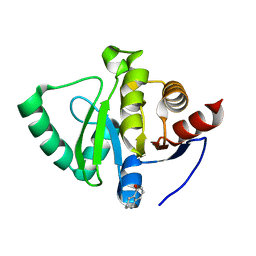 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with PB1827975385 | | Descriptor: | (5R)-5-amino-5,6,7,8-tetrahydronaphthalen-1-ol, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.037 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S2K
 
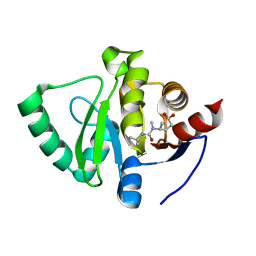 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z445856640 | | Descriptor: | N-[(3R)-1,1-dioxo-1lambda~6~-thiolan-3-yl]-N-methyl-N'-propan-2-ylurea, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S30
 
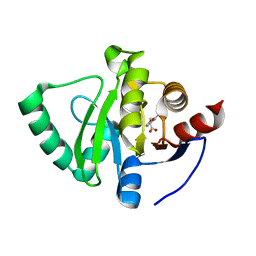 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z65532537 | | Descriptor: | (2R)-2-(2-fluorophenoxy)propanoic acid, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S3K
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z219104216 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 6-(ethylamino)pyridine-3-carbonitrile, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
