2GJE
 
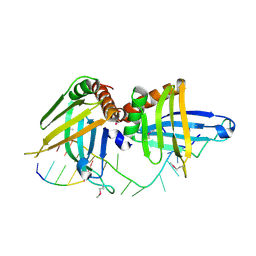 | | Structure of a guideRNA-binding protein complex bound to a gRNA | | Descriptor: | RNA tetramer, guide RNA 40-mer, mitochondrial RNA-binding protein 1, ... | | Authors: | Schumacher, M.A, Karamooz, E, Zikova, A, Trantirek, L, Lukes, J. | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Crystal Structures of T. brucei MRP1/MRP2 Guide-RNA Binding Complex Reveal RNA Matchmaking Mechanism.
Cell(Cambridge,Mass.), 126, 2006
|
|
1DBQ
 
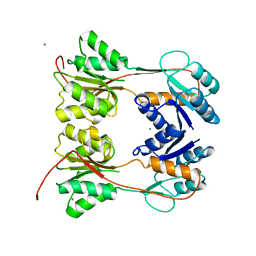 | | DNA-BINDING REGULATORY PROTEIN | | Descriptor: | MAGNESIUM ION, PURINE REPRESSOR | | Authors: | Schumacher, M.A, Choi, K.Y, Lu, F, Zalkin, H, Brennan, R.G. | | Deposit date: | 1996-02-13 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of corepressor-mediated specific DNA binding by the purine repressor.
Cell(Cambridge,Mass.), 83, 1995
|
|
4R22
 
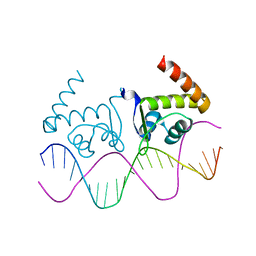 | | TnrA-DNA complex | | Descriptor: | DNA (5'-D(*CP*GP*TP*GP*TP*AP*AP*GP*GP*AP*AP*TP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), HTH-type transcriptional regulator TnrA | | Authors: | Schumacher, M.A. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
4R24
 
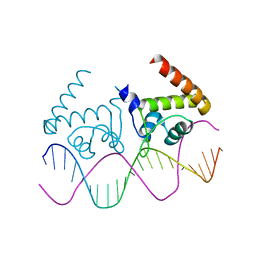 | | Complete dissection of B. subtilis nitrogen homeostatic circuitry | | Descriptor: | DNA (5'-D(*CP*GP*TP*GP*TP*AP*AP*GP*GP*AP*AP*TP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), HTH-type transcriptional regulator TnrA | | Authors: | Schumacher, M.A. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
4RX6
 
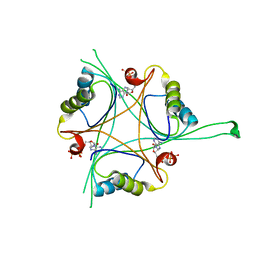 | | Structure of B. subtilis GlnK-ATP complex to 2.6 Angstrom | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Nitrogen regulatory PII-like protein | | Authors: | Schumacher, M.A, Cuthbert, B, Tonthat, N, Chinnam, N.G, Whitfill, T. | | Deposit date: | 2014-12-09 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5994 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
5TZG
 
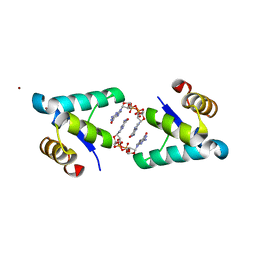 | | Structure of the BldD CTD(D116A)-(c-di-GMP)2, form 2 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein, ZINC ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex.
Nucleic Acids Res., 45, 2017
|
|
1Q88
 
 | |
2JPP
 
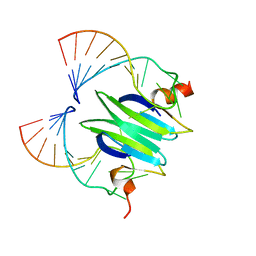 | | Structural basis of RsmA/CsrA RNA recognition: Structure of RsmE bound to the Shine-Dalgarno sequence of hcnA mRNA | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*UP*UP*CP*AP*CP*GP*GP*AP*UP*GP*AP*AP*GP*CP*CP*C)-3'), Translational repressor | | Authors: | Schubert, M, Lapouge, K, Duss, O, Oberstrass, F.C, Jelesarov, I, Haas, D, Allain, F.H.-T. | | Deposit date: | 2007-05-21 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA
Nat.Struct.Mol.Biol., 14, 2007
|
|
2NTZ
 
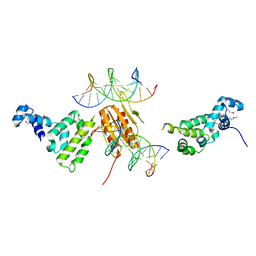 | |
2NZV
 
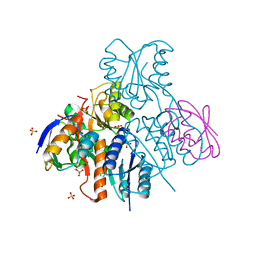 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors G6P and FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Catabolite control protein, Phosphocarrier protein HPr, ... | | Authors: | Schumacher, M.A, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-11-25 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
3EZ7
 
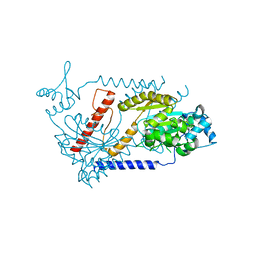 | |
3EZ9
 
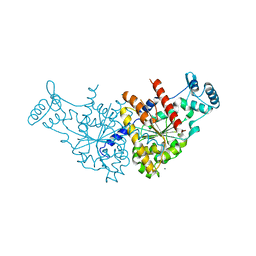 | | Partition Protein | | Descriptor: | MAGNESIUM ION, ParA | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA.
Embo J., 28, 2009
|
|
2OEN
 
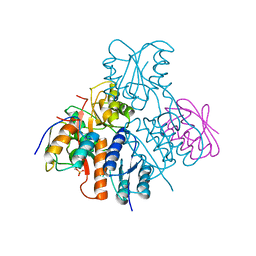 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors glucose-6-phosphate and fructose-1,6-bisphosphate | | Descriptor: | Catabolite control protein, Phosphocarrier protein HPr | | Authors: | Schumacher, M.A, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-12-30 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
3EZF
 
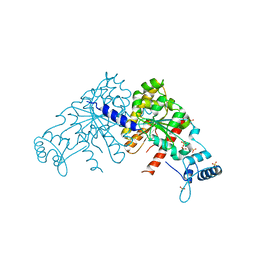 | | Partition Protein | | Descriptor: | ParA, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA.
Embo J., 28, 2009
|
|
3EZ6
 
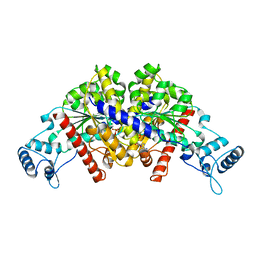 | | Structure of parA-ADP complex:tetragonal form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid partition protein A | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA.
Embo J., 28, 2009
|
|
1D4Z
 
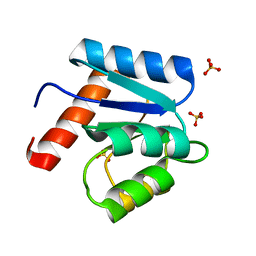 | | CRYSTAL STRUCTURE OF CHEY-95IV, A HYPERACTIVE CHEY MUTANT | | Descriptor: | CHEMOTAXIS PROTEIN CHEY, SULFATE ION | | Authors: | Schuster, M, Zhao, R, Bourret, R.B, Collins, E.J. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Correlated switch binding and signaling in bacterial chemotaxis.
J.Biol.Chem., 275, 2000
|
|
1ZX4
 
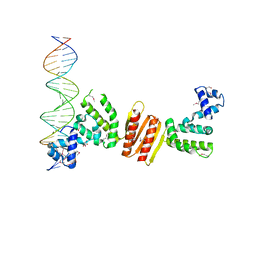 | | Structure of ParB bound to DNA | | Descriptor: | CITRIC ACID, Plasmid Partition par B protein, parS-small DNA centromere site | | Authors: | Schumacher, M.A, Funnell, B.E. | | Deposit date: | 2005-06-06 | | Release date: | 2005-11-29 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structures of ParB bound to DNA reveal mechanism of partition complex formation.
Nature, 438, 2005
|
|
5TZD
 
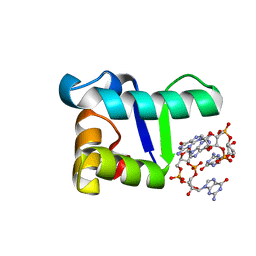 | | Structure of the WT S. venezulae BldD-(CTD-c-di-GMP)2 assembly intermediate | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | Authors: | Schumacher, M. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex.
Nucleic Acids Res., 45, 2017
|
|
1DBR
 
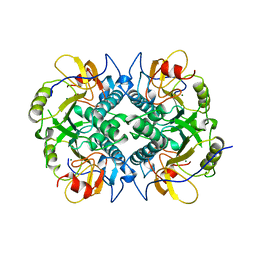 | | HYPOXANTHINE GUANINE XANTHINE | | Descriptor: | HYPOXANTHINE GUANINE XANTHINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION | | Authors: | Schumacher, M.A, Carter, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1996-02-13 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Toxoplasma gondii HGXPRTase reveal the catalytic role of a long flexible loop.
Nat.Struct.Biol., 3, 1996
|
|
3Q5W
 
 | |
5U1G
 
 | | Structure of TP228 ParA-AMPPNP-ParB complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ParA, TP228 ParB fragment | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structures of partition protein ParA with nonspecific DNA and ParB effector reveal molecular insights into principles governing Walker-box DNA segregation.
Genes Dev., 31, 2017
|
|
1Q87
 
 | |
1HAB
 
 | | CROSSLINKED HAEMOGLOBIN | | Descriptor: | 4-CARBOXYCINNAMIC ACID, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-03-13 | | Release date: | 1997-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric intermediates indicate R2 is the liganded hemoglobin end state.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1JUP
 
 | | Crystal structure of the multidrug binding transcriptional repressor QacR bound to malachite green | | Descriptor: | HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, MALACHITE GREEN, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-24 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
1HAC
 
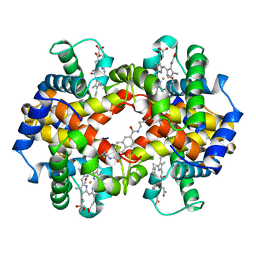 | | CROSSLINKED HAEMOGLOBIN | | Descriptor: | 2,6-DICARBOXYNAPHTHALENE, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-03-13 | | Release date: | 1997-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric intermediates indicate R2 is the liganded hemoglobin end state.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
