2VBG
 
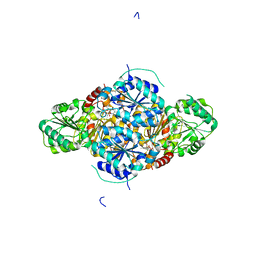 | | The complex structure of the branched-chain keto acid decarboxylase (KdcA) from Lactococcus lactis with 2R-1-hydroxyethyl-deazaThDP | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1R)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, BRANCHED-CHAIN ALPHA-KETOACID DECARBOXYLASE, MAGNESIUM ION | | Authors: | Berthold, C.L, Gocke, D, Wood, M.D, Leeper, F, Pohl, M, Schneider, G. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Branched-Chain Keto Acid Decarboxylase (Kdca) from Lactococcus Lactis Provides Insights Into the Structural Basis for the Chemo- and Enantioselective Carboligation Reaction
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2VHX
 
 | |
5AHK
 
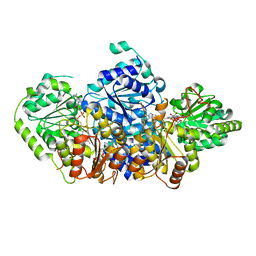 | | Crystal structure of acetohydroxy acid synthase Pf5 from Pseudomonas protegens | | Descriptor: | ACETOLACTATE SYNTHASE II, LARGE SUBUNIT, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Loschonsky, S, Mueller, M, Schneider, G. | | Deposit date: | 2015-02-06 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of the Acetohydroxy Acid Synthase Pf5 from Pseudomonas Protegens
To be Published
|
|
2WHR
 
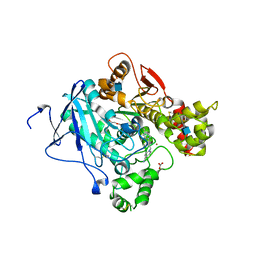 | | Crystal structure of acetylcholinesterase in complex with K027 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Ekstrom, F, Hornberg, A, Artursson, E, Hammarstrom, L.-G, Schneider, G, Pang, Y.-P. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Structure of Hi-6Sarin-Acetylcholinesterase Determined by X-Ray Crystallography and Molecular Dynamics Simulation: Reactivator Mechanism and Design.
Plos One, 4, 2009
|
|
2VHZ
 
 | |
2VHW
 
 | |
2WHP
 
 | | Crystal structure of acetylcholinesterase, phosphonylated by sarin and in complex with HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Ekstrom, F, Hornberg, A, Artursson, E, Hammarstrom, L.G, Schneider, G, Pang, Y.P. | | Deposit date: | 2009-05-06 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Hi-6Sarin-Acetylcholinesterase Determined by X-Ray Crystallography and Molecular Dynamics Simulation: Reactivator Mechanism and Design.
Plos One, 4, 2009
|
|
2WHQ
 
 | | Crystal structure of acetylcholinesterase, phosphonylated by sarin (aged) in complex with HI-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Ekstrom, F, Hornberg, A, Artursson, E, Hammarstrom, L.G, Schneider, G, Pang, Y.P. | | Deposit date: | 2009-05-06 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Hi-6Sarin-Acetylcholinesterase Determined by X-Ray Crystallography and Molecular Dynamics Simulation: Reactivator Mechanism and Design.
Plos One, 4, 2009
|
|
2F99
 
 | | Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity. | | Descriptor: | Aklanonic Acid methyl Ester Cyclase, AknH, SULFATE ION, ... | | Authors: | Kallio, P, Sultana, A, Neimi, J, Mantsala, P, Schneider, G. | | Deposit date: | 2005-12-05 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity.
J.Mol.Biol., 357, 2006
|
|
2GEX
 
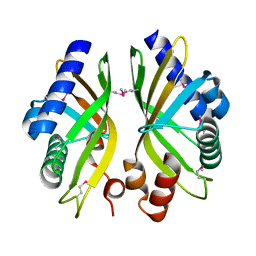 | |
3QZE
 
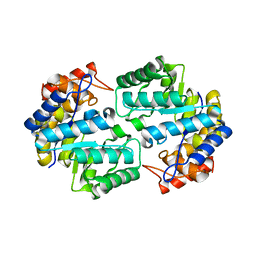 | |
2IPI
 
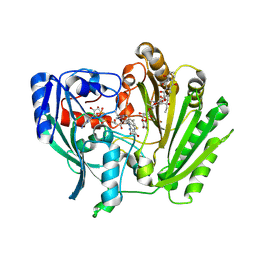 | | Crystal Structure of Aclacinomycin Oxidoreductase | | Descriptor: | Aclacinomycin oxidoreductase (AknOx), FLAVIN-ADENINE DINUCLEOTIDE, METHYL (2S,4R)-2-ETHYL-2,5,7-TRIHYDROXY-6,11-DIOXO-4-{[2,3,6-TRIDEOXY-4-O-{2,6-DIDEOXY-4-O-[(2S,6S)-6-METHYL-5-OXOTETRAHYDRO-2H-PYRAN-2-YL]-ALPHA-D-LYXO-HEXOPYRANOSYL}-3-(DIMETHYLAMINO)-D-RIBO-HEXOPYRANOSYL]OXY}-1,2,3,4,6,11-HEXAHYDROTETRACENE-1-CARBOXYLATE | | Authors: | Sultana, A, Kursula, I, Schneider, G, Alexeev, I, Niemi, J, Mantsala, P. | | Deposit date: | 2006-10-12 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure determination by multiwavelength anomalous diffraction of aclacinomycin oxidoreductase: indications of multidomain pseudomerohedral twinning.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3PBI
 
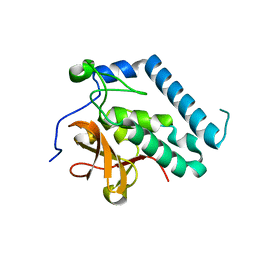 | |
5LB1
 
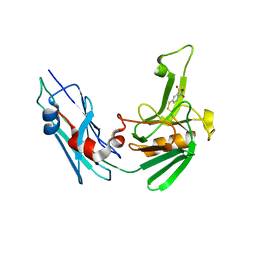 | | Crystal structure of the Mycobacterium tuberculosis L,D-transpeptidase-2 (LdtMt2) BC-module with thionitrobenzoate (TNB) adduct at the active site cysteine-354 | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, L,D-transpeptidase 2 | | Authors: | Steiner, E.M, Schnell, R, Schneider, G. | | Deposit date: | 2016-06-15 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Binding and processing of beta-lactam antibiotics by the transpeptidase LdtMt2 from Mycobacterium tuberculosis.
FEBS J., 284, 2017
|
|
5LBG
 
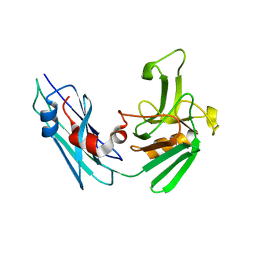 | | Crystal structure of the Mycobacterium tuberculosis L,D-transpeptidase-2 (LdtMt2) BC-module with faropenem-derived adduct at the active site cysteine-354 | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, L,D-transpeptidase 2 | | Authors: | Steiner, E.M, Schnell, R, Schneider, G. | | Deposit date: | 2016-06-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Binding and processing of beta-lactam antibiotics by the transpeptidase LdtMt2 from Mycobacterium tuberculosis.
FEBS J., 284, 2017
|
|
3PBC
 
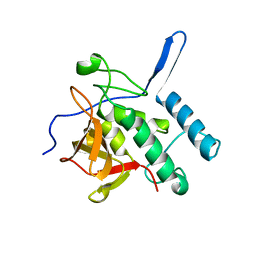 | |
3R5A
 
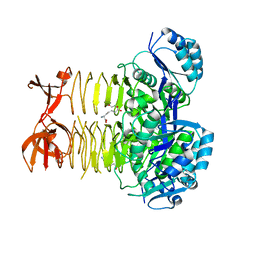 | |
3R5E
 
 | |
3R5C
 
 | |
3R5B
 
 | |
2GEY
 
 | | Crystal Structure of AclR a putative hydroxylase from Streptomyces galilaeus | | Descriptor: | AclR protein, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Beinker, P, Lohkamp, B, Schneider, G. | | Deposit date: | 2006-03-21 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of SnoaL2 and AclR: two putative hydroxylases in the biosynthesis of aromatic polyketide antibiotics
J.Mol.Biol., 359, 2006
|
|
3R5D
 
 | | Pseudomonas aeruginosa DapD (PA3666) apoprotein | | Descriptor: | GLYCEROL, Tetrahydrodipicolinate N-succinyletransferase | | Authors: | Sandalova, T, Schnell, R, Schneider, G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tetrahydrodipicolinate N-succinyltransferase and dihydrodipicolinate synthase from Pseudomonas aeruginosa: structure analysis and gene deletion.
Plos One, 7, 2012
|
|
2HP3
 
 | | Crystal structure of iminodisuccinate epimerase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IDS-epimerase, ... | | Authors: | Lohkamp, B, Bauerle, B, Rieger, P.G, Schneider, G. | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Three-dimensional Structure of Iminodisuccinate Epimerase Defines the Fold of the MmgE/PrpD Protein Family.
J.Mol.Biol., 362, 2006
|
|
3S0Q
 
 | |
2HP0
 
 | | Crystal structure of iminodisuccinate epimerase | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 1,2-ETHANEDIOL, IDS-epimerase, ... | | Authors: | Lohkamp, B, Bauerle, B, Rieger, P.G, Schneider, G. | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-12 | | Last modified: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional Structure of Iminodisuccinate Epimerase Defines the Fold of the MmgE/PrpD Protein Family.
J.Mol.Biol., 362, 2006
|
|
